| Review Article | ||
Open Vet. J.. 2021; 11(4): 569-580 Open Veterinary Journal, (2021), Vol. 11(4): 569–580 Review Article A recent perspective on fiber and hexon genes proteins analyses of fowl adenovirus toward virus infectivity—A reviewNorfitriah Mohamed Sohaimi1,2* and Mohd Hair-Bejo1,21Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Selangor, Malaysia 2Institute of Bioscience, Universiti Putra Malaysia, Serdang, Selangor, Malaysia *Corresponding Author: Norfitriah Mohamed Sohaimi. Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Selangor, Malaysia. Email: fitriahsohaimi [at] upm.edu.my Submitted: 17/07/2021 Accepted: 21/09/2021 Published: 19/10/2021 © 2021 Open Veterinary Journal
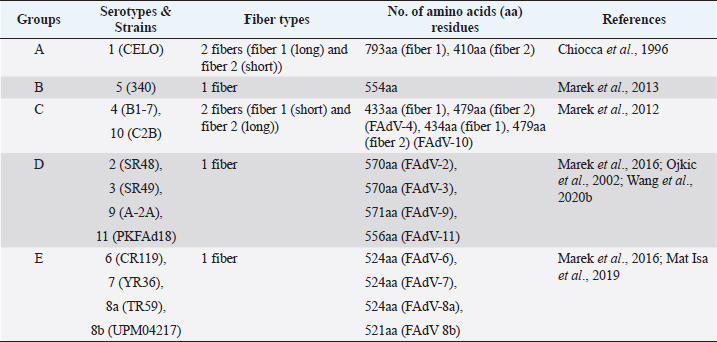
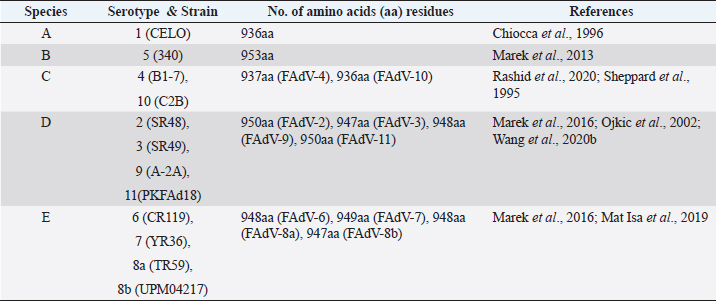
AbstractFowl adenovirus (FAdV) is a double-stranded DNA virus with a non-enveloped structure comprising three major proteins known as hexon, penton, and fiber. Molecular analysis which emphasizes on hexon and fiber proteins is currently the major focus of curiosity for FAdV antigenicity and pathogenicity. Recently, disease outbreaks associated with FAdV infections such as inclusion body hepatitis, hepatitis hydropericardium syndrome, and gizzard erosion, were commonly reported and continue to increase worldwide. Studies on the virulence gene of the virus were intensively conducted to provide a better understanding on the role of these major capsid proteins in the development of a safe and effective vaccine against the disease in the poultry industry. This paper highlights the variations of the fiber and hexon genes, their importance in genotypes and serotypes differentiation, and infectivity between FAdV strains. It appears that the L1 loop of hexon and the knob of fiber genes are the infectivity markers for FAdV infection. The fiber-2 protein plays a major role in FAdV pathogenicity than the hexon protein, while the fiber-1 protein is important for viral replication and assembly, regardless of virulence capability instead of infectivity. The hexon protein plays a major role in virus infectivity and tissue tropism. These findings could further enhance the knowledge of FAdV strains’ classification and evolution, diagnosis, and strategies to prevent and control FAdV infection and outbreaks in chicken farms. Keywords: Fowl adenovirus (FAdV), Infectivity, Hexon, Fiber, Pathogenicity. IntroductionIn the past two decades, studies on the variation of the fiber and hexon genes proteins among Fowl adenovirus (FAdV) species mainly focused on the antigenicity of the virus, but the role of these proteins on the infectivity in host cells remains scanty (Hess et al., 1995; Pallister et al., 1996; Hess, 2000). This is in contrast to human adenovirus (AdV), where studies on both major capsid proteins were comprehensive, involving various genotypes and serotypes with a clear pathway of viral mechanisms prior to infection (Walters et al., 2002; Varghese et al., 2004; Kalyuzhniy et al., 2008; Russell, 2009). Recently, investigation of the protein genes encoded for FAdV virulence determinant and infectivity were of major interest to many researchers worldwide toward the development of a safe and effective vaccine against the disease (Okuda et al., 2006; Park et al., 2017; Schachner et al., 2018; Sohaimi et al., 2019; Pan et al., 2020; Wang et al., 2020a). It was suggested that fiber and hexon genes play a major role as a virulence determinant of FAdV infection (Pallister et al., 1996; Sohaimi et al., 2018). Analysis on both major capsid proteins was attempted in various studies as reported previously (Park et al., 2017; Yasmeen et al., 2017; Sohaimi et al., 2019). It is the intention of this paper to review the available literature on the analysis of fiber and hexon genes which highlighted the clinical diseases associated with FAdV, classification, and purposes of these proteins. Clinical diseases caused by FAdV infectionFowl adenovirus infections are a major threat to the poultry industry and cause several clinical diseases in chickens with a significant economic impact due to mortality and poor productivity (Morshed et al., 2017; Norfitriah et al., 2018; Cizmecigil et al., 2020; Cui et al., 2020). Recently, disease outbreaks associated with FAdV infections, such as inclusion body hepatitis (IBH), hepatitis hydropericardium syndrome (HHS), and gizzard erosion, were commonly reported and continue to increase worldwide. Occasionally, FAdV has been reported as a causative agent in cases of necrotizing pancreatitis and respiratory disease in chickens (Dhillon et al., 1982; Tanimura et al., 1993). IBH was first reported in 1963 in USA (Helmboldt and Frazier, 1963). Since then, the disease has spread globally mainly in meat-producing chickens of 2–3 weeks old and some in layer chickens of 25–27 weeks old (Norina et al., 2016; Norfitriah et al., 2018; Abghour et al., 2019; Jordan et al., 2019). Epidemiological studies confirmed that IBH is commonly caused by either FAdV serotypes 2 or 8a, 8b, and 11 (Morshed et al., 2017; Schachner et al., 2018). IBH was characterized by the sudden onset of high mortality which peaked after 3–4 days postinfection (pi) and returned to normal on day 5 but occasionally continued for 2–3 weeks (Hair-Bejo, 2005). Mortality is relatively low; however, the sick chickens exhibit clinical signs of depression, ruffled feathers, and reduced feed consumption prior to death (Hafez, 2011). IBH can affect the entire poultry production, such as in broiler, layer, and breeder chickens. Mortality may reach 10% and occasionally it is higher up to 30% (Gomis et al., 2006). Gross lesions of IBH are characterized by hepatic necrosis and inflammation with friable, swollen, pale, and petechial hemorrhages in the liver of the affected chickens (Morshed et al., 2017). Histologically, numerous basophilic and eosinophilic intranuclear inclusion bodies (INIBs) are mainly found in degenerated hepatocytes either surrounded by a clear halo or filling with an entire enlarged nucleus (Hair-Bejo, 2005). HHS is caused by FAdV serotype 4 and is characterized by sudden death in broiler chickens with high mortality ranging from 20% to 80% (Ahmad et al., 2011; Mahmood et al., 2014). The disease was first reported in 1987 in Pakistan (Khawaja et al., 1988) and subsequently in various geographical regions such as India, South Korea, and China (Kim et al., 2008; Kataria et al., 2013; Ren et al., 2019). It mainly affects commercial broiler flocks of 3–5 weeks old with lesions of a swollen pericardial sac filled with straw-colored fluid, and swollen and friable livers (Ahmad et al., 1989; Anjum et al., 1989). INIBs were observed in the liver, while extensive congestion and hemorrhages were recorded in the heart (Ren et al., 2019). In addition, severe depletion of lymphocytes in the bursa of Fabricius, thymus, and spleen caused by the high pathogenic strain from FAdV serotype 4 was recorded (Hussain et al., 2012; Niu et al., 2019). The disease was also reported in breeder and laying flocks with low mortality of below 10% compared to broiler chickens (Balamurugan and Kataria, 2004). HHS causes huge economic losses to the poultry industry, especially due to the high mortality rate (Balamurugan and Kataria, 2004; Zhang et al., 2016). Adenoviral gizzard erosion has been continuously reported in many countries due to FAdV serotypes 1 and 8b in broiler and layer chickens (Ono et al., 2003; Schachner et al., 2018; Norfitriah et al., 2018; Mirzazadeh et al., 2021). The disease affects the broiler flock’s performance with a reduction in body weight gain, high mortality of up to 80%, and increased condemnation rate at the slaughterhouse (Schade et al., 2013; Mirzazadeh et al., 2021). A drop in egg production with malformation of eggs was observed in layer chickens infected with the virus (Norfitriah et al., 2018). Discoloration and erosion of koilin layer and dilated proventriculus and gizzard were commonly reported. The gizzard mucosa was necrotic with the presence of intranuclear inclusion bodies in the degenerating glandular epithelial cells (Ono et al., 2003; Mirzazadeh et al., 2021). FAdV classification by L1 loop of hexon gene and fiber gene proteinsClassification of FAdV is based on the highly variable region of the L1 loop in hexon gene and a lesser extent in the fiber gene of the virus. The genotyping of FAdV is designated as species A–E and divided further into 12 serotypes (Steer et al., 2011; Schachner et al., 2016). Each species consist of one or more serotypes as follows: FAdV-1 is the only member in species A; FAdV-5 is a unique member for the species B; FAdV-4 and FAdV-10 are members in species FAdV-C; FAdV-2, FAdV-3, FAdV-9, and FAdV-11 are members of species FAdV-D; and FAdV-6, FAdV-7, FAdV-8a, and FAdV-8b are members within species FAdV-E (Kaján et al., 2013; Marek et al., 2013). Fiber as capsid projector protein of FAdVThe fiber protein is a projection from the virus capsid which consists of an N-terminal tail inserted into the penton base, a thin shaft consisting of variable length depending on the serotype of FAdVs, and a carboxyl-terminal knob or head domain containing the receptor-binding sites (Mathis et al., 2005; Grgić et al., 2011). It is about 2 nm thick with lengths between 10 nm and 47 nm depending on FAdVs serotypes (Gelderblom and Maichle-Lauppe, 1982). Two fiber proteins with different lengths as short and long fibers were identified in FAdV species A, which are attached to a single penton base (Hess et al., 1995; Chiocca et al., 1996). In the CELO strain (serotype 1), the short and long fiber lengths are 42.5 and 8.5 nm, respectively, binding to different receptors on the cell surface, permitting virus attachment and internalization (Hess et al., 1995; Chiocca et al., 1996). In addition, two fibers with equal length are exclusively found in FAdV species C (Marek et al., 2012). On the contrary, only one fiber is present in other species of FAdV such as in species B (serotype 5), D (serotypes 2, 3, 9, and 12), and E (serotypes 6, 7, 8a, and 8b) (Grgić et al., 2014; Marek et al., 2016). The length of amino acid residues varies based on serotypes of FAdV, as summarized in Table 1. High amino acid variability encoded in fiber protein mainly at a region of head domain or knob results in binding to different receptors (Hess et al., 1995). In the knob region of the fiber protein, large fractions of the antigenic site exist in all serotypes and possess a type-specific epitope for neutralization with the antibody (Sheppard and Trist, 1992). Hexon as a major capsid proteinThe hexon protein constitutes a large proportion of adenovirus capsid comprising 240 non-vertex capsomeres which form homotrimer structures that give rise to a pseudohexagonal shape with a triangular top superimposed on the base. The sizes of hexon molecules vary between species and serotypes of FAdVs (Russell, 2009). The length of amino acids’ residues varies between FAdV serotypes, as summarized in Table 2 (Sheppard et al., 1995; Chiocca et al., 1996; Ojkic et al., 2002; Marek et al., 2013; Mat Isa et al., 2019; Rashid et al., 2020). Table 1. Amino acid residues of fiber protein among fowl adenovirus (FAdV) species and serotypes.
Table 2. Amino acid residues of hexon protein among fowl adenovirus (FAdV) species and serotypes.
The structure of the hexon protein is divided into conserved pedestal regions located inside the virion (P1 and P2) and variable loops exposed on the outer surfaces between serotypes to from type-specific epitopes, namely L1, L2, and L4 (Roberts et al., 1986; Crawford-Miksza and Schnurr, 1996). The L3 loop is buried internally and is more conserved to stabilize the interface between the P1 and P2 conserved regions (Roberts et al., 1986; Toogood and Hay, 1988). Most of the amino acid variations such as mutation, deletion, or additions are frequently found in the three intertwined loops due to high immunological selective pressures for neutralization with antibodies (Roberts et al., 1986; Toogood and Hay, 1988). Analysis of hypervariable regions (HVRs) in hexon gene from L1 loop comprised HVRs 1–4 with different lengths for each region (Niczyporuk, 2018). They are 191 bp long in HVR1, 50 bp long in HVR2, 90 bp long in HVR3, and 18 bp long in HVR4 (Niczyporuk, 2018). There are major differences between FAdVs types encoded in the sequence of HVRs; however, they are constant for every species. Moreover, the L1 loop regions are the highest sequence variability and longest loop in protein with a complicated folding structure which serves as the location of specific receptors (Gelderblom and Maichle-Lauppe, 1982; Niczyporuk, 2018). Mutations in HVRs caused adenovirus to circumvent the host immunity mechanisms since these regions are responsible for antibodies’ binding (Niczyporuk, 2018). In addition, sequences encoded in HVRs reflect high differentiation between species or types and between the strains which infect different hosts. Analysis of the HVR1 region revealed that the amino sequence is specific for the exact adenovirus host (Rux et al., 2003). Based on previous work, five conservative sequences were identified in HVRs as described by Niczyporuk (2018) as follows: GQMTN, GQMTT, GQLSN, GQMTH, and GQMS. The GQMT sequence at the end of the HVR1 was observed specifically for types FAdV-1, FAdV-2 and 11, FAdV-5, and FAdV-7 and 8b. For strains from FAdV-4 and -8a, the sequences encoded as GQLS and GQMS, respectively (Niczyporuk, 2018). HVR1 form the hairpin structure and is found in all FAdV types as the main site for neutralizing antibody binding. Recently, the fiber gene was the protein highlighted as a gene encoding for virulence instead of the hexon gene due to its structure protruding from the surface (Wang et al., 2018; Pallister et al., 1996). To date, fiber and hexon proteins have become a major focus of research interest in determination of tissue tropism and virulence of the virus (Pallister et al., 1996; Park et al., 2017; Sohaimi et al., 2018; Zhang et al., 2018). It is vital to understand the roles of the hexon and fiber genes in the infectivity of the virus for effective prevention and control of the disease, especially for the development of safe and effective vaccines. Divergence of fiber gene protein sequences between FAdV strainsUtilization of fiber gene sequences for molecular differentiation between FAdV species was emphasized in previous works with variations in intraspecies diversity (Marek et al., 2012; Liu et al., 2016; Shachner et al., 2016; Sohaimi et al., 2019). Analysis of predicted fiber protein sequences showed that the N-terminal tail was highly conserved between IBH field isolates with 63 residues for all FAdV-D and FAdV-E fibers with only 3 non-synonymous substitutions in FAdV-D strains, while 2 non-synonymous among FAdV-E strains (Shachner et al., 2016). In cases of IBH, both FAdV-D and FAdV-E strains were commonly isolated in poultry species worldwide and were highlighted intensively in previous works (Kaján et al., 2013; Zadravec et al., 2013; Marek et al., 2016; Kaján et al., 2019). Tentative nuclear localization signal (RKRP) at position aa17-aa20 is conserved with the exception of FAdV-D (RKRPàRKRS). Similarly, for the FAdV-E fiber tail sequence, the penton base interaction motif at aa53–aa56 exhibited motif alteration from VYPF à VHPF. When compared to the shaft region, the sequence is more conserved as demonstrated between FAdV-D field strains and positioned at the middle of the fiber protein (Shachner et al., 2016). Interestingly, huge non-synonymous substitutions in the head domain or knob region were detected and resulted in the divergence of two separate amino acid patterns mainly in FAdV-E field isolates (Schahner et al., 2016). These differences are crucial to determine the specific host cell receptor since this region is the primary site for virus attachment prior to infection (Hess et al., 1995). Among the FAdV-E strains, there is greater variability in the fiber protein when compared to hexon genes which are consistent with human adenoviruses (La Rosa et al., 2011). On the contrary, the degree of variability in the two proteins was relatively equivalent between FAdV-D strains (Shachner et al., 2016). Comparative sequence analysis between FAdV-1, CELO strain, and human adenovirus (HAdV) subgenus F, type Ad40 from Dugan strain, revealed only 25% identity based on short and long fiber gene proteins (Chiocca et al., 1996). The sequence of the non-pathogenic CELO strain indicates a viral genome of 43.8 kb, which is 9 kb longer than the 34.2kb genome of HAdV (Chiocca et al., 1996). Major differences in the fiber protein between the adenovirus genera were recorded, although both investigated strains consist of two fibers and bind to coxsackievirus and adenovirus receptor (CAR) prior to infection. In addition, the differences in virus infectivity were also noted since AdV40 from HAdV causes acute gastroenteritis in humans, while the CELO strain is non-pathogenic in chickens (Chiocca et al., 1996). The analysis of fiber protein between FAdV-1 and Atadenovirus genus from egg drop syndrome (EDS) outbreak revealed only 32% identity and the findings confirmed the serological distinctness between two avian adenovirus genera in chickens (Hess et al., 1997). Phylogenetic analyses between FAdV (Aviadenovirus) and other avian adenovirus genera, Siadenovirus from cases of turkey hemorrhagic enteritis and Atadenovirus (EDS), indicate large divergences among avian adenovirus genera based on fiber genes sequences, regardless of virus virulence in chicken (Sohaimi et al., 2019). Although some strains of FAdV are highly pathogenic in chickens, similar to turkey hemorrhagic enteritis (HE) and EDS in chickens or ducks, it seems that there are major variations in this protein, which are clustered into a different branch in the phylogenetic tree (Sohaimi et al., 2019). Fiber gene variation between pathogenic and non-pathogenic FAdV strainsIt was reported that the differences in virulence of FAdV between CFA40 and CFA3 strains were due to variation of gene encoded in the knob region of the fiber gene (Pallister et al., 1996). Similarly, in other studies, the amino acid identities in the fiber gene between IBH and non-IBH strains from FAdV serotype 8 were low with only 89% along with a total of 22 non-synonymous mutations (Grgić et al., 2014). A study conducted by Mase et al. (2010) revealed that there were 14 amino acid substitutions among FAdV-4 strains between HHS strains in short fiber protein. These changes in amino acid residues of a single fiber gene were critical for tissue tropism and virulence of FAdV strains inducing IBH in chickens (Wang et al., 2014; Mei and Wadell, 1995). Recent studies confirmed that the fiber-2 gene from the FAdV-4 strain was closely associated with the virulence and pathogenicity of the virus (Zhang et al., 2018). Analysis on the short fiber gene between pathogenic and non-pathogenic FAdV-1 strains, PL/G060/08 and CELO, respectively, showed 13 different nucleotides which led to the substitution of amino acid residues at N223K, I314T, R328G, F331G, S334A, and A369C in the knob region (Domanska-Blicharz et al. 2011). When compared to the long fiber gene, 18 nucleotides’ differences were noted between PL/G060/08 and CELO with only 1 amino acids substitution at T275A in the shaft region. It showed that variations in amino acids are prominent in the short fiber protein which is crucial for infection of chicken cells as described by Tan et al. (2001). As a result, the FAdV-1 PL/G060/08 strain caused 100% mortality and gizzard erosion in SPF chickens (Domanska-Blicharz et al. 2011). Fiber-2 and hexon proteins encoded for virulence determinant between FAdV strainsGenomic identification and characterization of the FAdV strains on fiber and hexon genes provide valuable information and are critically important for prevention and control strategies against the disease. It was observed that the FAdV-4 strain from China, SDSX1, comprised 479aa residues in which 5aa residues were more than non-pathogenic strain, ON1, in the fiber-2 gene (Li et al., 2018). The basic residues’ rich sequences KRPK/KRAK (site 27–30) and VYPF (site 41–45) in fiber proteins of SDSX1 were identified. More than 20 substitutions of aa were detected in the isolate compared to the non-pathogenic strains, ON1 and KR5, with low identity based on the sequence analysis. The strain was highly pathogenic in chickens and caused 100% mortality. Analysis of the fiber-2 gene indicated that protein substitutions may respond to the pathogenicity of the strain (Li et al., 2018). Genetic characterization of the pathogenic strain from FAdV-4, namely MX-SHP95 with other HHS strains and non-pathogenic strains, KR5 and ON1, revealed amino acid substitutions at I188R in the hexon protein, S432G in fiber-1, and highest in fiber-2 protein at G219D, I300T, S305A, P307A, I378T, A380T, T435S, and S453A, as reported by Liu et al. (2016). These findings are consistent with a recent study conducted by Rashid et al. (2020). Multiple amino acid substitutions were also noticed with 12aa differences in the fiber-2 region and minimal variation in fiber-1 between HHS strains and non-HHS strains (Rashid et al., 2020). Moreover, multiple sequence alignments between strains resulted in aa substitutions at G219D, P307A, V319I, and A380T within fiber-2 which is conserved among FAdV-4 isolates from HHS cases compared to the non-pathogenic isolates (Marek et al., 2012; Liu et al., 2016). This suggests that fiber-2 serves as one primary virulence factor gene strain since HHS strains are clustered together in the phylogenetic tree with close evolutionary relationship, although they are from different regions worldwide and are similar to non-pathogenic strains (Liu et al., 2016). Molecular analysis of amino acid sequences of hexon and fiber-2 by multiple sequence alignment revealed several changes between highly virulent strain, HNJZ, and non-pathogenic strain, ON1. These changes were observed in the L1 loop of the hexon protein as follows: 164 S, 188 R, 193 R, 195 Q, 238 D, 240 T, 243 N, 263 I, and 264 V. In addition, 10 aa changes were noticed in the fiber-2 protein as follows: 219 D, 232 Q, 300 T, 305 A, 307 A, 329 L, 378 T, 380 T, 435 S, and 453 A. Comparison of the terminal tail region showed that fiber-2 of the HNJZ strain consist of 479aa residues, in which 5 aa was more than ON1 strain at position 11–15 aa ENGKP (Zhang et al., 2018). Based on earlier literature, the knob region in the fiber protein is involved in virus attachment to the host cell (Pallister et al., 1996). This suggests that changes in amino acids in regions of tail and knob resulted in different binding affinities toward host cell receptors and subsequently virus infectivity in chickens (Wang et al., 2014). Genetic modification of fiber-2 and hexon genes from the pathogenic strain FAdV-4 caused the development of clinical signs associated with HHS in chickens which were similar to natural infections (Zhang et al., 2018). Fiber-2 and hexon play a crucial role in the pathogenicity of FAdV. Based on virulence testing, the recombinant viruses comprising fiber-2 and hexon genes play important role in virulence determinants between the highly virulent strain and non-pathogenic strain. Chickens infected with mutant FAdV-4 fiber-2 isolate caused 100% mortality when compared to mutant FAdV-4 hexon isolate which caused only 50% mortality throughout the trial. Differences in mortality pattern indicate that fiber-2 plays a major role in FAdV-4 pathogenicity than hexon protein. A study on the non-pathogenic strain of FAdV-4, ON1 strain, revealed neither clinical signs nor death in the inoculated chickens (Zhang et al., 2018). Furthermore, analysis of the fiber-2 gene revealed amino acid variations between HHS and non-HHS isolates at D219 and T300, which were conserved for HHS isolates from five countries compared to non-HHS isolates as reported previously (Park et al., 2017). These changes in aa residues are consistent with earlier findings which are possibly relevant to virulence (Mase et al., 2010; Marek et al., 2012; Vera-Hernández et al., 2016; Park et al., 2017). As reported in previous studies, differences in the knob domain of the fiber gene as well as in the L1 loop of the hexon genes were involved in the differences in tissue tropism and virulence for human and canine adenoviruses (Mei and Wadell, 1995; Rasmussen et al., 1995; Sohaimi et al., 2018). The hexon protein constitutes a large proportion of virion and more conserved among FAdV strains compared to fiber protein which protrudes from the surface. In a recent study, the substitution of amino acid residue was detected among pathogenic FAdV-4 strains and unique to these strains at position 188 (I–R) in the hexon protein (Rashid et al., 2020). According to a study conducted by Zhang et al. (2018), it was confirmed that hexon proteins from the FAdV-4 strain are closely associated with the virulence and pathogenicity of the virus (Zhang et al., 2018). A study conducted by Dar et al. (2012) revealed that FAdV isolates obtained from the IBH outbreak, IBHV(SK)m were highly identical to FAdV-8b strain 764 with 99.5% in the L1 loop of hexon gene. The isolate caused high mortality in young chicks following experimental infection (Dar et al., 2012). A recent study found that FAdV-4 strain obtained from China in cases of HHS was pathogenic to SPF chickens with mild to severe HHS lesions. The isolate was closely related to other common local strains based on hexon gene analysis (Cui et al., 2020). Amino acid variations encoded in hexon gene proteins contribute to a significant impact toward pathogenicity in chickens. According to Niczyporuk and Czekaj (2016), the pathogenic strain FAdV-1/A-61/11z exhibited 17 substitutions and 5 deletions of amino acids in the L1 loop compared to apathogenic strain, FAdV-8a/E-6/12j, with only 3 different amino acids. Pathogenicity testing indicated that the strain with prominent molecular changes in the L1 loop of hexon protein caused mortality and lesions in infected SPF chickens (Niczyporuk and Czekaj, 2016). In addition, sequence analysis of FAdV-4 strains indicated the lowest identity about 47.2% than other serotypes based on HVR1 regions in the L1 loop (Niczyporuk and Czekaj, 2016). The finding was confirmed by the data presented in 2007 by Pichla-Gollon et al. (2007) where the side of the hairpin structure of HVR1 is the main site for the neutralizing antibody binding. As a result, the mutation in this region could trigger FAdV to circumvent host immunity, and perhaps the main factor for most virulent FAdV-4 strains caused HHS with high mortality and severe lesions in chickens (Niczyporuk and Czekaj, 2016). Implication of FAdV isolates passaging in alternative mediums toward proteins and virus infectivityIt is well noted that hexon protein plays a major role in virus infectivity and tissue tropism in human Adv since it constitutes a large proportion of virus capsid (Kalyuzhniy et al., 2008; Short et al., 2010). A similar role of the hexon gene for encoding virulence determinant may also occur for FAdV infection in poultry. Recent studies have demonstrated that the consecutive passages of FAdV isolates in alternative mediums either in cell cultures or chicken embryonated eggs (CEE) resulted in modification of hexon and fiber gene proteins. It was proved that at the 20th and 16th passages of the FAdV isolates in CEE, it caused molecular changes in both major capsid proteins which encoded for virulence determinant and attenuation of the isolates (Pallister et., 1996; Mansoor et al., 2011; Sohaimi et al., 2018). This finding is in corroborated with previous works in cell cultures (Ali et al., 2015; Sohaimi et al., 2019). To maintain viability in SPF CEE for 20th passage, substitution in hexon and fiber proteins is critical for the continuous growth of the FAdV isolate. It appears that nucleotide changes at T90C in the L1 loop and knob region of fiber gene at G1078C with amino acid substitutions at A360P are consistent from early to passage 20 (Sohaimi et al., 2018). Similarly, for the FAdV passages in chicken embryo liver (CEL) cells, changes at T90C were also detected at 35th passage as a marker for adaptation and attenuation in this cell line. Moreover, this marker gene was not detected in the original isolate before virus passages (Sohaimi et al., 2019). Comparative sequence analysis before and after attenuation produced several nucleotide and amino acids residue changes at different positions based on the medium used to attenuate the virus isolate. In CEE, 4nt bases were substituted at T90C, A147G, C199T, and A1134T, resulting in the substitution of amino acid residues at T49A, A66V, and M378L in hexon gene at passage 20 isolates (Sohaimi et al., 2018). On the contrary, the molecular changes in similar passage numbers (CEL20) in CEL cells were minimal with only 2nt substitutions, resulting in a change of 1aa due to the rapid formation of CPE which take only 24–48 hours post-inoculation (pi) and an insufficient period for hexon protein modification by the virus. Therefore, the changes were prominent at passage 35 (CEL35) with 4nt substitutions at T90C, A133T, C400T, and T556A in the L1 loop which caused the substitution of amino acid residues at D44V, S133F, and V185E (Sohaimi et al., 2019). A study conducted using FAdV-4 isolate revealed that the virulent field strain was attenuated only at the 16th passage probably because the virus had adequate time to alter the hexon structure with lesions produced which takes up to 120 hours pi. These changes were noted with 2% and 3% differences in nucleotides and amino acid sequences, respectively, between before and after attenuation in the variable L1 loop region (Mansoor et al., 2011). Analysis of the fiber gene revealed gene modification in the passaged isolate from passage 20 (E20) in SPF chicken embryonated eggs (CEEs) with nucleotides and amino acid substitutions at A879G, T918C, C952G, A964T, and A1062C and in amino acid residues at P318A and N322Y within the knob region (Sohaimi et al., 2018). Similarly, the adaptation and attenuation of FAdV isolate in CEL cells necessitate an increased number of passages until 35 times compared to CEEs. Several nucleotide changes were noticed at T556C, T821A, A1042C, and A1062C, resulting in non-synonymous changes in amino acid residues at L189P and F274Y in the shaft region, followed by T348P and A360P in the knob region (Sohaimi et al., 2019) The impact of the hexon and fiber genes changes were attempted in CEEs with delayed mortality pattern of embryos and non-visible lesions at a higher passage (Mansoor et al., 2011; Sohaimi et al., 2018). It was consistent in CEL cells at passage 35 of the isolate with delayed in cytopathic effect (CPE) formation and non-pathogenic in SPF chickens (Sohaimi et al., 2019). It is interesting to note that the attenuated isolates induced antibody responses in chickens at day 21 post-inoculation (Mansoor et al., 2011; Sohaimi et al., 2019). It appears that molecular changes in the L1 loop of hexon and the knob of fiber genes are infectivity markers for FAdV (Sohaimi et al., 2018). Short fiber (fiber-1) of FAdV-4 as a key mediator for infectionThe fiber proteins play vital roles in the FAdV infection and pathogenesis as demonstrated by recent studies (Sohaimi et al., 2019; Liu et al., 2020; Wang et al., 2020a). It was confirmed that fiber-2 is encoded as a virulence determinant, whereas fiber-1 is a key factor for directly mediating the infection of FAdV-4 through its shaft and knob domains according to molecular analysis and pathogenicity findings in chickens. In addition, fiber-1 and its knob domain may serve as a target for identifying the receptor for FAdV-4 (Wang et al., 2020a). Moreover, superinfection resistance analysis and an interfering assay indicated that fiber-1 triggering FAdV-4 infection instead of fiber-2. Following truncation analysis, both shaft and knob domains of fiber-1 were required for the infection. The sera against the knob domain were able to block FAdV-4 infection and the knob-containing fusion protein provided efficient protection against the lethal challenge of FAdV-4 in chickens (Wang et al., 2020a). It was supported by other studies that fiber-1 genes are necessary for virus replication, regardless of virulence capability (Liu et al., 2020). Interaction of FAdV fiber with the cellular receptor in the host is the primary step in human AdV infection. The application of protein blockage and antibody-neutralization assays capable of confirming the function of short fiber was critical for binding to susceptible leghorn male hepatocellular cells (Pan et al., 2020). Different serotypes possess different structures of fiber proteins. As shown, FAdV-1 and FAdV-4 comprised two fibers, long and short length, based on several amino acid residues. The mechanism of FAdV binding to the receptor is distinct between FAdV-1 and FAdV-4. Although both serotypes are made up of two fibers, the length and function for each fiber are significantly different. Fiber-1 of the FAdV-4 was recognized as a shorter fiber than fiber-2 based on the number of amino acids (aa) and protein size in the virus. Short fiber is critically used for binding to surface molecules on host cells prior to infection (Pan et al., 2020). Compared to human AdV, the interaction between knob domain and CAR has a weak binding affinity and is unlikely to utilize this molecule as a primary receptor in vivo (Baker et al., 2019). Reverse genetic system clarified fiber-1 roles for FAdV replication and assemblyThe fiber-1 mutant virus was constructed based on the FAdV-4 infectious clone of hypervirulent strain HNJZ using Redαβ recombineering techniques, followed by pathogenicity in SPF chickens (Liu et al., 2020). A reverse genetic system using the fiber-1 gene from hypervirulent strain, HNJZ, was replaced with fiber-1 gene obtained in non-pathogenic strain ON1. It appears that that mutant virus caused 100% mortality in SPF chickens with similar HHS gross and histopathological changes, which indicated that the role of fiber-1 is important for viral replication and assembly instead of infectivity. The study conducted by Liu et al. (2020) revealed that the mutant virus with fiber-1 gene ON1 and hypervirulent strain HNJZ had a higher viral load compared to non-pathogenic ON1 which suggested that fiber-1 gene, regardless of virulence capability, are essential for virus replication. The fiber-1 gene involved in replication instead of virulence determinant since both strains caused 100% mortality in SPF chickens (Liu et al., 2020). Other studies demonstrated that fiber-1 protein induces immune protection against the hypervirulent strain of FAdV-4 and is crucial for binding with cellular receptor CAR prior to infection (Wang et al., 2018; Pan et al., 2020). FAdV-1 long fiber (fiber-1) for attachment to CAR receptorPrevious studies have revealed that the long fiber (fiber-1) of FAdV-1 is essential for binding to the CAR to facilitate virus entry into the host for infection. On the contrary, the short fiber (fiber-2) was critical for infection of chicken cells (Tan et al., 2001; Taharaguchi et al., 2012). It seems that analysis of the long fiber genes by polymerase chain reaction-restriction fragment length polymorphism pattern between pathogenic and non-pathogenic strains distinguished the pathogenicity and strains of FAdV-1. The nucleotide sequences were distinct between strains that induced gizzard erosion, 99ZH, and the one without induced any clinical disease, Ote strain (Ono et al., 2004; Okuda et al., 2006). The isolates obtained from the gizzard erosion outbreak produced similar restriction patterns as those pathogenic strains; 99ZH was compared to isolates from clinically healthy chickens which was similar to the Ote strain. The RFLP findings were compatible with the pathogenicity trial in SPF chickens. All the isolates with similar patterns to 99ZH strain induced gizzard erosion in SPF chickens, while not in other isolates which were similar to the Ote strain (Ono et al., 2003, 2004). Utilization of the RFLP assay was capable of differentiating between strains and confirmed pathogenicity trial in chickens (Okuda et al., 2006). Fiber-2 and hexon genes’ vaccines from FAdV-4 are immunogenic in chickensRecombinant vaccines comprising fiber-1, fiber-2, and hexon-L1 loop were generated for efficacy testing against virulent FAdV-4 (Schachner et al., 2014; Wang et al., 2018). It showed that the fiber-2 vaccine conferred higher protection than fiber-1 and hexon-L1 loop vaccines in the vaccinated chickens. The recombinant fiber-2 was recognized as a protective immunogen since the vaccine was capable of inducing a high antibody response gradually after vaccination and potentially used as a subunit vaccine candidate to prevent HHS in chickens (Schachner et al., 2014). These findings are compatible with a study conducted by Wang et al. (2018). It seems that the subunit vaccine comprised fiber-2 gene which induced excellent protection against FAdV-4 challenge, followed by fiber-1, hexon, and penton. When the vaccine dose increased, the protections induced by fiber-1, hexon, and penton increased, indicating a dose-dependent relationship between these vaccines and their level of immune protection against FAdV-4. Additionally, hexon induces immune response as subunit vaccines and confers full protection against FAdV-4 infection when compared to penton protein (Wang et al., 2018). Cross-reactive activities of the fiber monoclonal antibodiesThe sandwich ELISA assay demonstrated that monoclonal antibodies targeting fiber-1 FAdV-4 were cross-reactive with fiber-1 FAdV-10 due to similar genotype species and highly homologous based on fiber protein sequences (Shao et al., 2019). Moreover, monoclonal antibodies developed by Lu et al. (2019) specific to fiber protein FAdV-8 recognize the common epitope in the fiber proteins between FAdV-7 and FAdV-8. ConclusionThe fiber and hexon proteins encoded virulence determinants for FAdV and play a critical role in virus infectivity. It appears that molecular changes in the L1 loop of hexon and the knob of fiber genes are the infectivity markers for FAdV. The fiber-2 protein plays a major role in FAdV pathogenicity than the hexon protein, while the fiber-1 protein is important for viral replication and assembly, regardless of virulence capability instead of infectivity. The hexon protein plays a major role in virus infectivity and tissue tropism. Analyses of both proteins contribute to broader knowledge on viral evolution between FAdV strains and strategies to prevent and control the occurrence of FAdV infection and disease outbreaks in poultry farms. Conflict of interestThe authors declare that there is no conflict of interest. Authors’ contributionsNMS wrote the article and reviewed the previous papers. MHB wrote and edited the final draft of the manuscript. All authors read and approved the final manuscript. ReferencesAbghour, S., Zro, K., Mouahid, M., Tahiri, F., Tarta, M., Berrada, J. and Kichou, F. 2019. Isolation and characterization of fowl aviadenovirus serotype 11 from chickens with inclusion body hepatitis in Morocco. PLoS One. 14(12), e0227004 Ahmad, I., Afzal, M., Malik, M.I., Hussain, Z. and Hanif, W. 1989. Studies on the disease pattern and etiology of hydropericardium syndrome (Angara disease) in broiler chickens in Pakistan. Pak. J. Agric. Res. 10, 195–199. Ahmad, M.D., Zaman, S., Mushtaq, M.H., Anjum, A.A. and Akram, M. 2011. Comparative pathogenicity of liver homogenate and cell culture propagated hydropericardium syndrome virus in broiler birds. Pak. Vet. J. 31(4), 321–326. Anjum, A., Sabri, M., and Iqbal, Z. 1989. Hydropericarditis syndrome in broiler chickens in Pakistan. Vet. Record. 124, 247–248. Baker, A.T., Greenshields-Watson, A., Coughlan, L., Davies, J.A., Uusi-Kerttula, H., Cole, D.K., Rizkallah, P.J. and Parker, A.L. 2019. Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions. Nat. Commun. 10(1), 741. Balamurugan, V. and Kataria, J.M. 2004. The hydropericardium syndrome in poultry- a current scenario. Vet. Res. Commun. 28, 127–148. Chiocca, S., Kurzbauer, R., Schaffner, G., Baker, A., Mautner, V. and Cotten, M. 1996. The complete DNA sequence and genomic organization of the avian adenovirus CELO. J. Virol. 70(5), 2939–2949. Cizmecigil, U. Y., Umar, S., Yilmaz, A., Bayraktar, E., Turan, N., Tali, B., Aydin, O., Tali, H. E., Yaramanoglu, M., Yilmaz, S. G., Kolukisa, A., Sadeyen, J. R., Iqbal, M. and Yilmaz, H. (2020). Characterisation of Fowl Adenovirus (FAdV-8b) Strain Concerning the Geographic Analysis and Pathological Lesions Associated with Inclusion Body Hepatitis in Broiler Flocks in Turkey. J. Vet. Res. 64(2), 231–237. Cui. J., Xu, Y., Zhou, Z., Xu, Q., Wang, J., Xiao, Y., Li, Z. and Bi, D. 2020. Pathogenicity and molecular typing of Fowl Adenovirus-associated associated with hepatitis/hydropericardium syndrome in Central China (2015–2018). Front. Vet. Sci. 7, 190. Crawford-Miksza, L. and Schnurr, D.P. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70(3), 1836–1844. Chiocca, S., Kurzbauer, R., Schaffner, G., Baker, A. and Mautner, V. 1996. The complete DNA sequence and genomic organization of the avian adenovirus CELO. J. Virol. 70, 2939–2949. Dar, A., Gomis, S., Shirley, I., Mutwiri, G., Brownlie, R., Potter, A., Gerdts, V. and Tikoo, SK. 2012. Pathotypic and molecular characterization of a fowl adenovirus associated with inclusion body hepatitis in Saskatchewan chickens. Avian Dis. 56(1), 73-81. Dhillon, A.S., Winterfield, R.W., Thacker, H.L. and Feldman, D.S. 1982. Lesions induced in the respiratory tract of chickens by serologically different adenoviruses. Avian Dis. 26, 478–486. Domanska-Blicharz, K., Tomczyk, G., Smietanka, K., Kozaczynski, W. and Minta, Z. 2011. Molecular characterization of fowl adenoviruses isolated from chickens with gizzard erosions. Poult. Sci. 90(5), 983–989. Gelderblom H, Maichle-Lauppe I. 1982. The fibers of fowl adenoviruses. Arch. Virol. 72(4), 289–298. Gomis, S., Goodhope, R., Ojkić, D. and Wilson, P. 2006. Inclusion body hepatitis as a primary disease in broilers in Saskatchewan, Canada. Avian Dis. 50(4), 550–555. Grgić, H., Krell, P.J. and Nagy, E. 2014. Comparison of fiber gene sequences of inclusion body hepatitis (IBH) and non-IBH strains of serotype 8 and 11 fowl adenoviruses. Virus Genes 48(1), 74–80. Grgić, H., Yang, D-H., Nagy, E. 2011. Pathogenicity and complete genome sequence of fowl adenovirus serotype 8 isolate. Virus Res. 156(1-2), 91–97. Hafez, H.M. 2011. Avian adenovirus infections with special attention to inclusion body hepatitis/hydropericardium syndrome and egg drop syndrome. Pak. Vet. J. 31, 85–92. Hair-Bejo, M. 2005. Inclusion body hepatitis in commercial broiler chickens. J. Vet. Malaysia 17, 23–26. Helmboldt, C.F. and Frazier, M.N. 1963. Avian hepatic inclusion bodies of unknown significance. Avian Dis. 7, 446–450. Hess, M. 2000. Detection and differentiation of avian adenoviruses: A review. Avian Pathol. 29(3), 195–206. Hess, M., Blöcker, H. and Brandt, P. 1997. The complete nucleotide sequence of the egg drop syndrome virus: an intermediate between mastadenoviruses and aviadenoviruses. Virology 238(1), 145–156. Hess, M., Cuzange, A., Ruigrok, R.W., Chroboczek, J. and Jacrot, B. 1995. The avian adenovirus penton: two fibers and one base. J. Mol. Biol. 252(4), 379–385. Hussain, I., Mahmood, M.S., Arshad, M.I., Akhtar, M., Mahmood, F. and Rafique, A. 2012. Immune system dysfunction in broiler chickens experimentally inoculated with fowl adenovirus serotype-4 associated with inclusion body hepatitis hydropericardium syndrome. Turkish J. Vet. Anim. Sci. 36(3), 223–230. Jordan, A.B., Blake, L., Bisnath, J., Ramgattie, C., Carrington, C.V. and Oura, C.A.L. 2019. Identification of four serotypes of fowl adenovirus in clinically affected commercial poultry co-infected with chicken infectious anaemia virus in Trinidad and Tobago. Transbound. Emerg. Dis. 66(3), 1341–14348. Khawaja, D.A., Ahmad, S., Rauf, A.M., Zulfiqar, M., Mahmood, S.M. and Hassan, M. 1988. Isolation of an adenovirus from hydropericardium syndrome in broiler chicks. Pak. J. Vet. Res. 1, 2–27. Kaján, G.L., Affranio, I., Tóthné-Bistyák, A., Kecskeméti, S. and Benkő, M. 2019. An emerging new fowl adenovirus genotype. Heliyon. 5(5), e01732. Kaján, G.L., Kecskeméti, S., Harrach, B. and Benkő, M. 2013. Molecular typing of fowl adenoviruses, isolated in Hungary recently, reveals high diversity. Vet. Microbiol. 167(3–4), 357–363. Kalyuzhniy, O., Di Paulo, N.C., Silvestry, M., Hofherr, S.E., Barry, M.A., Stewart, P.L. and Shayakhmetov, D.M. 2008. Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc. Natl. Acad. Sci. USA. 105, 5483–5488. Kataria, J.M., Dhama, K., Nagarajan, S., Chakraborty, S., Kaushal, A. and Deb, R. 2013. Fowl adenoviruses causing hydropericardium syndrome in poultry. Adv. Anim. Vet. Sci. 1(4S), 5–13. Kim, J. N., Byun, S. H., Kim, M. J., Kim, J. J., Sung, H. W. and Mo, I. P. 2008. Outbreaks of hydropericardium syndrome and molecular characterization of Korean fowl adenoviral isolates. Avian Dis. 52(3), 526–530. La Rosa, G., Iaconelli, M., Pourshaban, M., Luca, E., Valentini, P., Sica, S., Manzara, S., Delogu, G. and Muscillo, M. 2011. Molecular characterization of adenovirus from clinical samples through analysis of the hexon and fiber genes. J. Gen. Virol. 92, 412–420. Li, L., Wang, J., Chen, P., Zhang, S., Sun, J. and Yuan, W. 2018. Pathogenicity and molecular characterization of a fowl adenovirus 4 isolated from chicken associated with IBH and HPS in China. BMC Vet. Res. 14(1), 400. Liu, R., Zhang, Y., Guo, H., Li, N., Wang, B., Tian, K., Wang, Z., Yang, X., Li, Y., Wang, H., Zhang, Y., Fu, J. and Zhao, J. 2020. The increased virulence of hypervirulent fowl adenovirus 4 is independent of fiber-1 and penton. Research in Veterinary Science. 131, 31–37. Liu, Y., Wan, W., Gao, D., Li, Y., Yang, X., Liu, H., Yao, H., Chen, L., Wang, C. and Zhao, J. 2016. Genetic characterization of novel fowl aviadenovirus 4 isolates from outbreaks of hepatitis-hydropericardium syndrome in broiler chickens in China. Emerg. Microb. Infect. 5(11), e117. Lu, H., Shao, H., Chen, H., Zhang, J., Wang, W., Li, T., Xie, Q., Qin, A. and Ye, J. 2019. Identification of novel B cell epitopes in the fiber protein of serotype 8 Fowl adenovirus. AMB Express 9(1), 172. Mahmood, M.S., Ali, S., Hussain, I., Aslam, A. and Rafique, A. 2014. The development of hydropericardium syndrome vaccines. World`s Poult. Sci. J. 70(2), 355–364. Mansoor, M.K., Hussain, I., Arshad, M. and Muhammad, G. 2011. Preparation and evaluation of chicken embryo-adapted fowl adenovirus serotype 4 vaccine in broiler chickens. Trop. Anim. Health Prod. 43(2), 331–338. Mat Isa, N., Mohd Ayob, J., Ravi, S., Mustapha, N.A., Ashari, K.S., Bejo, M.H., Omar, A.R. and Ideris, A. 2019. Complete genome sequence of fowl adenovirus-8b UPM04217 isolate associated with the inclusion body hepatitis disease in commercial broiler chickens in Malaysia reveals intermediate evolution. Virus Dis. 30(3), 426–432. Marek, A., Nolte, V., Schachner, A., Berger, E., Schlötterer, C. and Hess, M. 2012. Two fiber genes of nearly equal lengths are a common and distinctive feature of Fowl adenovirus C members. Vet. Microbiol. 156(3–4), 411–417. Marek, A., Kosiol, C., Harrach, B., Kaján, G.L., Schlötterer, C. and Hess, M. 2013. The first whole genome sequence of a fowl adenovirus B strain enables interspecies comparisons within the genus Aviadenovirus. Vet. Microbiol. 166, 250–256. Marek, A., Kaján, G.L., Kosiol, C., Benkő, M., Schachner, A. and Hess, M. 2016. Genetic diversity of species Fowl aviadenovirus D and Fowl aviadenovirus E. J. Gen. Virol. 97(9), 2323–2332. Mase, M., Nakamura, K. and Imada, T. 2010. Characterization of Fowl adenovirus serotype 4 isolated from chickens with hydropericardium syndrome based on analysis of the short fiber protein gene. J. Vet. Diagn. Invest. 22(2), 218–223. Mathis, J.M., Stoff-Khalili, M.A. and Curiel, D.T. 2005. Oncolytic adenoviruses—selective retargeting to tumor cells. Oncogene 24(52), 7775–7791. Mirzazadeh, A., Grafl, B., Abbasnia, M., Emadi-Jamali, S., Abdi-Hachesoo, B., Schachner, A. and Hess, M. 2021. Reduced Performance Due to Adenoviral Gizzard Erosion in 16-Day-Old Commercial Broiler Chickens in Iran, Confirmed Experimentally. Front. Vet. Sci. 8, 635186. Mei, Y.F. and Wadell, G. 1995. Molecular determinants of adenovirus tropism. Curr. Top. Microbiol. Immunol. 199, 213–228. Morshed, R., Hosseini, H., Langeroudi, A.G., Fard, M.H.B. and Charkhkar, S. 2017. Fowl Adenoviruses D and E Cause Inclusion Body Hepatitis Outbreaks in Broiler and Broiler Breeder Pullet Flocks. Avian Dis. 61(2), 205–210. Niczyporuk, J.S. 2018. Deep analysis of Loop L1 HVRs1-4 region of the hexon gene of adenovirus field strains isolated in Poland. PLoS One. 13(11), e0207668. Niczyporuk, J. S. and Czekaj, H. 2018. A comparative pathogenicity analysis of two adenovirus strains, 1/A and 8a/E, isolated from poultry in Poland. Arch. Virol. 163(11), 3005–3013. Niu, Y., Sun, Q., Shi, Y., Ding, Y., Li, Z., Sun, Y., Li, M. and Liu, S. 2019. Immunosuppressive potential of fowl adenovirus serotype 4. Poult. Sci. 98(9), 3514–3522. Norfitriah, M.S., Hair-Bejo, M., Omar, A.R., Aini, I. and Nurulfiza, M.I. 2018. Molecular detection and pathogenicity of fowl adenovirus isolated from disease outbreak in commercial layer chickens. Int. J. Agric. Sci. Vet. Med. 6(1), 73–84. Norina, L., Norsharina, A., Nurnadiah, A.H., Redzuan, I., Ardy, A. and Nor-Ismaliza, A. 2016. Avian adenovirus isolated from broiler affected with inclusion body hepatitis. Malays. J. Vet. Res. 7(2), 121–126. Ojkic, D., Krell, P.J. and Nagy, E. 2002. Unique features of fowl adenovirus 9 gene transcription. Virology 302(2), 274–285. Okuda, Y., Ono, M., Shibata, I., Sato, S. and Akashi, H. 2006. Comparison of the polymerase chain reaction-restriction fragment length polymorphism pattern of the fiber gene and pathogenicity of serotype-1 fowl adenovirus isolates from gizzard erosions and from feces of clinically healthy chickens in Japan. J. Vet. Diagn. Invest. 18(2), 162–167. Ono, M., Okuda, Y., Yazawa, S., Shibata, I., Sato, S. and Okada, K. 2003. Outbreaks of adenoviral gizzard erosion in slaughtered broiler chickens in Japan. Vet. Rec. 153, 775–779. Ono, M., Okuda, Y., Yazawa, S., Imai, Y., Shibata, I., Sato, S. and Okada, K. 2003. Adenoviral gizzard erosion in commercial broiler chickens. Vet. Pathol. 40(3), 294–303. Ono, M., Okuda, Y., Shibata, I., Sato, S. and Okada, K. 2004. Pathogenicity by parenteral injection of fowl adenovirus isolated from gizzard erosion and resistance to reinfection in adenoviral gizzard erosion in chickens. Vet. Pathol. 41(5), 483–489. Pan, Q., Wang, J., Gao, Y., Wang, Q., Cui, H., Liu, C., Qi, X., Zhang, Y., Wang, Y., Li, K., Gao, L., Liu, A. and Wang, X. 2020. Identification of chicken CAR homology as a cellular receptor for the emerging highly pathogenic fowl adenovirus 4 via unique binding mechanism. Emerg. Microb. Infect. 9, 586–596. Park, H.S., Lim, I.S., Kim, S.K., Kim, T.K., Park, C.K. and Yeo, S.G. 2017. Molecular analysis of the hexon, penton base, and fiber-2 genes of Korean fowl adenovirus serotype 4 isolates from hydropericardium syndrome-affected chickens. Virus Genes 53(1), 111–116. Pallister, J., Wright, P.J. and Sheppard, M. 1996. A single gene encoding the fiber is responsible for variations in virulence in the fowl adenoviruses. J. Virol. 70(8), 5115–5122. Pichla-Gollon, S.L., Drinker, M., Zhou, X., Xue, F., Rux, J.J., Gao, G.P., Wilson, J.M., Ertl, H.C., Burnett, R.M. and Bergelson, J.M. 2007. Structure-based identification of a major neutralizing site in an adenovirus hexon. J. Virol. 81(4), 1680–1689. Rashid, F., Xie, Z., Zhang, L., Luan, Y., Luo, S., Deng, X., Xie, L., Xie, Z. and Fan, Q. 2020. Genetic characterization of fowl aviadenovirus 4 isolates from Guangxi, China, during 2017-2019. Poult. Sci. 99(9), 4166–4173. Rasmussen, U.B., Schlesinger, Y., Pavirani, A. and Mehtali, M. 1995. Sequence analysis of the canine adenovirus 2 fiber-encoding gene. Gene 159, 279–280. Ren, G., Wang, H., Yan, Y., Liu, F., Huang, M. and Chen, R. 2019. Pathogenicity of a fowl adenovirus serotype 4 isolated from chickens associated with hydropericardium-hepatitis syndrome in China. Poult. Sci. 98(7), 2765–2771. Roberts, M.M., White, J.L., Grütter, M.G. and Burnett, R.M. 1986. Three-dimensional structure of the adenovirus major coat protein hexon. Science 232(4754), 1148–1151. Russell, W.C. 2009. Adenoviruses: update on structure and function. J. Gen. Virol. 90(1), 1–20. Rux, J.J., Kuser, P.R. and Burnett, R.M. 2003. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution x-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol. 77, 9553–9566. Schachner, A., Matos, M., Grafl, B. and Hess, M. 2018. Fowl adenovirus-induced diseases and strategies for their control—a review on the current global situation. Avian Pathol. 47(2), 111–126. Schachner, A., Marek, A., Grafl, B. and Hess, M. 2016. Detailed molecular analyses of the hexon loop-1 and fibers of fowl aviadenoviruses reveal new insights into the antigenic relationship and confirm that specific genotypes are involved in field outbreaks of inclusion body hepatitis. Vet. Microbiol. 186, 13–20. Schachner, A., Marek, A., Jaskulska, B., Bilic, I. and Hess, M. 2014. Recombinant FAdV-4 fiber-2 protein protects chickens against hepatitis-hydropericardium syndrome (HHS). Vaccine 32(9), 1086–1092. Schade, B., Schmitt, F., Bohm, B., Alex, M., Fux, R., Cattoli, G., Terregino, C., Monne, I., Currie, R.J. and Olias, P. 2013. Adenoviral gizzard erosion in broiler chickens in Germany. Avian Dis. 57(1), 159–163. Shao, H., Lu, Y., Wang, W., Li, T., Zhang, J., Wan, Z., Liang, G., Gao, W., Qin, A. and Ye, J. 2019. Two novel monoclonal antibodies against fiber-1 protein of FAdV-4 and their application in detection of FAdV-4/10. BMC Vet. Res. 15, 232. Sheppard, M., McCoy, R.J. and Werner, W. 1995. Genomic mapping and sequence analysis of the fowl adenovirus serotype 10 hexon gene. J. Gen. Virol. 76(10), 2595–2600. Sheppard, M. and Trist, H. 1992. Characterization of the avian adenovirus penton base. Virology 188(2), 881–886. Short, J.J., Rivera, A.A., Wu, H., Walter, M.R., Yamamoto, M., Mathis, J.M. and Curiel, D.T. 2010. Substitution of adenovirus serotype 3 hexon onto a serotype 5 oncolytic adenovirus reduces factor X binding, decreases liver tropism, and improves antitumor efficacy. Mol. Cancer Ther. 9(9), 2536–2544. Sohaimi, N.M., Bejo, M.H., Omar, A.R., Ideris, A. and Isa, N.M. 2019. Molecular characterisation of fowl adenovirus isolate of Malaysia attenuated in chicken embryo liver cells and its pathogenicity and immunogenicity in chickens. PLoS One. 14(12), e0225863. Sohaimi, N.M., Bejo, M.H., Omar, A.R., Ideris, A. and Isa, N.M. 2018. Hexon and fiber gene changes in an attenuated fowl adenovirus isolate from Malaysia in embryonated chicken eggs and its infectivity in chickens. J. Vet. Sci. 19(6), 759–770. Steer, P.A., O'Rourke, D., Ghorashi, S.A. and Noormohammadi, A.H. 2011. Application of high-resolution melting curve analysis for typing of fowl adenoviruses in field cases of inclusion body hepatitis. Aust. Vet. J. 89(5), 184–192. Taharaguchi, S., Fukazawa, R., Kitazume, M., Harima, H., Taira, K., Oonaka, K. and Hara, M. 2012. Biology of fowl adenovirus type 1 infection of heterologous cells. Arch. Virol. 157(11), 2223–2226. Tan, P.K., Michou, A.I., Bergelson, J.M. and Cotton, M. 2001. Defining CAR as a cellular receptor for the avian adenovirus CELO using a genetic analysis of the two viral fibre proteins. J. Gen. Virol. 82(6), 1465–1472. Tanimura, N., Nakamura, K., Imai, K., Maeda, M., Gobo, T., Nitta, S., Ishihara, T. and Amano, H. 1993. Necrotizing pancreatitis and gizzard erosion associated with adenovirus infection in chickens. Avian Dis. 37(2), 606–611. Toogood, C.I. and Hay, R.T. 1988. DNA sequence of the adenovirus type 41 hexon gene and predicted structure of the protein. J. Gen. Virol. 69(9), 2291–2301. Varghese, R., Mikyas, Y., Stewart, P.L. and Ralston, R. 2004. Postentry neutralization of adenovirus type 5 by an antihexon antibody. J. Virol. 78, 12320–12332. Vera-Hernández, P.F., Morales-Garzón, A., Cortés-Espinosa, D.V., Galiote-Flores, A., García-Barrera, L.J., Rodríguez-Galindo, E.T., Toscano-Contreras, A., Lucio-Decanini, E. and Absalón, A.E. 2016. Clinicopathological characterization and genomic sequence differences observed in a highly virulent fowl Aviadenovirus serotype 4. Avian Pathol. 45(1), 73–81. Walters, R.W., Freimuth, P., Moninger, T.O., Ganske, I., Zabner, J. and Welsh, M.J. 2002. Adenovirus fiber disrupts CAR-mediated intercellular adhesion allowing virus escape. Cell 110, 789–799. Wang, W., Liu, Q., Li, T., Geng, T., Chen, H., Xie, Q., Shao, H., Wan, Z., Qin, A. and Ye, J. 2020a. Fiber-1, Not Fiber-2, Directly Mediates the Infection of the Pathogenic Serotype 4 Fowl Adenovirus via Its Shaft and Knob Domains. J. Virol. 94(17), e00954-20. Wang, J., Zaheer, I., Saleemi, M.K., Qi, X., Gao, Y., Cui, H., Li, K., Gao, L., Fayyaz, A., Hussain, A., Liu, C., Zhang, Y., Wang, X. and Pan, Q. 2020b. The first complete genome sequence and pathogenicity characterization of fowl adenovirus 11 from chickens with inclusion body hepatitis in Pakistan. Vet. Microbiol. 244, 108670. Wang, X., Tang, Q., Chu, Z., Wang, P., Luo, C., Zhang, Y., Fang, X., Qiu, L., Dang, R. and Yang, Z. 2018. Immune protection efficacy of FAdV-4 surface proteins fiber-1, fiber-2, hexon and penton base. Virus Res. 245, 1–6. Wang Z, Wang B, Lou J, Yan J, Gao L, Geng R, Yu B. 2014. Mutation in fiber of adenovirus serotype 5 gene therapy vector decreases liver tropism. Int. J. Clin. Exp. Med. 7(12), 4942–4950. Yasmeen, S., Siddique, N., Athar Abbas, M., Ali, A., Rafique, S., Rashid, F., Shah, A.U., Mehmood, F., Begum, I., Javaid, T., Jaffery, S., Ali, R. and Naeem, K. 2017. Fiber gene based molecular and biological characterization of hydropericardium-hepatitis syndrome associated avian adenoviruses. Iran. J. Vet. Res. 18(3), 190–196. Zadravec, M., Slavec, B., Krapez, U., Kaján, G.L., Racnik, J., Juntes, P., Jursic-Cizerl, R., Benkö, M. and Zorman Rojs, O. 2013. Inclusion body hepatitis (IBH) outbreak associated with fowl adenovirus type 8b in broilers. Acta Vet. 63, 101–110. Zhang, Y., Liu, R., Tian, K., Wang, Z., Yang, X., Gao, D., Zhang, Y., Fu, J., Wang, H. and Zhao, J. 2018. Fiber2 and hexon genes are closely associated with the virulence of the emerging and highly pathogenic fowl adenovirus 4. Emerg. Microb. Infect. 7(1), 199. | ||
| How to Cite this Article |
| Pubmed Style Sohaimi NM, Bejo MH. A recent perspective on fiber and hexon genes proteins analyses of fowl adenovirus towards virus infectivity - A review. Open Vet. J.. 2021; 11(4): 569-580. doi:10.5455/OVJ.2021.v11.i4.6 Web Style Sohaimi NM, Bejo MH. A recent perspective on fiber and hexon genes proteins analyses of fowl adenovirus towards virus infectivity - A review. https://www.openveterinaryjournal.com/?mno=98642 [Access: January 25, 2026]. doi:10.5455/OVJ.2021.v11.i4.6 AMA (American Medical Association) Style Sohaimi NM, Bejo MH. A recent perspective on fiber and hexon genes proteins analyses of fowl adenovirus towards virus infectivity - A review. Open Vet. J.. 2021; 11(4): 569-580. doi:10.5455/OVJ.2021.v11.i4.6 Vancouver/ICMJE Style Sohaimi NM, Bejo MH. A recent perspective on fiber and hexon genes proteins analyses of fowl adenovirus towards virus infectivity - A review. Open Vet. J.. (2021), [cited January 25, 2026]; 11(4): 569-580. doi:10.5455/OVJ.2021.v11.i4.6 Harvard Style Sohaimi, N. M. & Bejo, . M. H. (2021) A recent perspective on fiber and hexon genes proteins analyses of fowl adenovirus towards virus infectivity - A review. Open Vet. J., 11 (4), 569-580. doi:10.5455/OVJ.2021.v11.i4.6 Turabian Style Sohaimi, Norfitriah Mohamed, and Mohd Hair Bejo. 2021. A recent perspective on fiber and hexon genes proteins analyses of fowl adenovirus towards virus infectivity - A review. Open Veterinary Journal, 11 (4), 569-580. doi:10.5455/OVJ.2021.v11.i4.6 Chicago Style Sohaimi, Norfitriah Mohamed, and Mohd Hair Bejo. "A recent perspective on fiber and hexon genes proteins analyses of fowl adenovirus towards virus infectivity - A review." Open Veterinary Journal 11 (2021), 569-580. doi:10.5455/OVJ.2021.v11.i4.6 MLA (The Modern Language Association) Style Sohaimi, Norfitriah Mohamed, and Mohd Hair Bejo. "A recent perspective on fiber and hexon genes proteins analyses of fowl adenovirus towards virus infectivity - A review." Open Veterinary Journal 11.4 (2021), 569-580. Print. doi:10.5455/OVJ.2021.v11.i4.6 APA (American Psychological Association) Style Sohaimi, N. M. & Bejo, . M. H. (2021) A recent perspective on fiber and hexon genes proteins analyses of fowl adenovirus towards virus infectivity - A review. Open Veterinary Journal, 11 (4), 569-580. doi:10.5455/OVJ.2021.v11.i4.6 |