| Research Article | ||
Open Veterinary Journal, (2024), Vol. 14(1): 242-255 Original Research Moringa oleifera as a potential antimicrobial against pathogenic Clostridium perfringens isolates in farm animalsGhada A. Ibrahim1* and Khadijah A. Altammar21Agriculture Research Center (ARC), Animal Health Research Institute (AHRI), Bacteriology Department, Ismailia Branch, Egypt 2Department of Biology, College of Science, University of Hafr Al Batin, Hafr Al Batin, Saudi Arabia *Corresponding Author: Ghada A. Ibrahim. Bacteriology Department, Agriculture Research Center (ARC), Animal Health Research Institute (AHRI), Giza, Egypt. Email: Dr.ghadaabdelaal [at] ahri.gov.eg Submitted: 01/10/2023 Accepted: 15/12/2023 Published: 31/01/2024 © 2024 Open Veterinary Journal
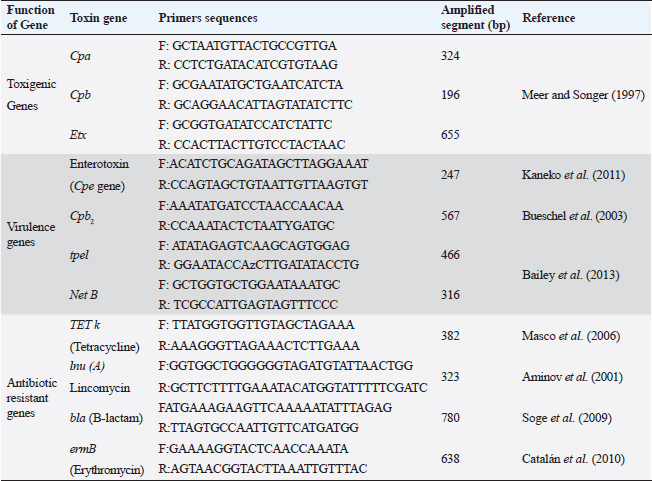
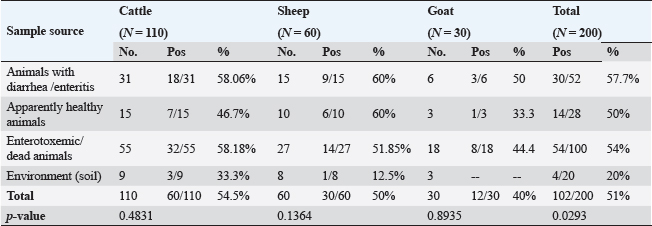
ABSTRACTBackground: Clostridium perfringens (CP) is an emerging anaerobic pathogen that can aggravate severe fatal infections in different hosts and livestock. Aim: This paper was designed to monitor the antibacterial efficacy of Moringa oleifera (M. oleifera) plant against different CP isolates of variant toxin genotypes comparing that with commercial antibiotics in the veterinary field. Methods: A total of 200 examined fecal, intestinal, and liver samples from cattle, sheep, and goats were investigated bacteriologically and biochemically for CP. Then, the isolates were examined by polymerase chain reaction (PCR) for toxin gene typing. Thereafter, the antimicrobial susceptibility testing as well as the antibacterial efficacy of M. oleifera were evaluated and statistically analyzed against recovered isolates. Results: The prevalence rate of CP was 51% (102/200); of which 54.5% was from cattle, 50% from sheep, and 40% from goat. Moreover, all CP isolates were highly resistant to tetracycline and lincomycin drugs; meanwhile, they were of the least resistance against ciprofloxacin (8.3%–16.7%), cefotaxime (16.7%–25%), and gentamycin (26.7%–33.3%). For M. oleifera, high antibacterial efficacy with greater inhibition zones of the plant was recorded with its oil (20–24 mm) and ethanolic extracts (16–20 mm) against CP than the aqueous extract (≤ 10 mm). A good correlation was stated between M. oleifera oil and toxin type of CP isolates particularly type A followed by D and B types. Interestingly, the oil and ethanolic extracts of M. oleifera gave higher antibacterial efficacy than most commercial antibiotics against the recovered isolates. Conclusion: This study highlighted the potent antibacterial properties of M. oleifera for suppressing CP isolated from farm animals; hence, more investigations on M. oleifera are suggested to support its use as a medical herbal plant substituting antibiotics hazards and resistance problems worldwide. Keywords: Antibacterial efficacy, C. perfringens, Genotyping, Moringa oleifera, PCR. IntroductionClostridium perfringens (CP) are anaerobic pathogens that have been accused of variable diseases and syndromes in most livestock animals (Nazki et al., 2017). CP organisms are Gram-positive, ubiquitous, spore-forming, and non motile rods (0.6–0.8×2-4 μm). They are commonly inhabitant of the gastrointestinal tract of humans and animals. Their pathogenicity has been mediated by the ability to produce a wide range of toxins, which could strongly impact animal clinical signs, severity of disease, and enteric infections in different animal species (Ohtani and Shimizu, 2016). Based on the produced toxin, CP is classified into five types from A–E (CPA, CPB, CPC, CPD, and CPE). These pathogens release mainly alpha, beta, epsilon, and iota toxins (ITX) (Milton et al., 2017). However, the alpha toxin is the major released toxin in all five types of CP; CPB produces three types of toxins: alpha, beta, and epsilon (ETX toxin) and CPC produces both alpha and beta toxins. Moreover, CPD strains produce epsilon along with alpha toxin and CPE strains produce alpha and ITX. CP strains of type A toxin were involved in enteritis, severe intestinal problems, food poisoning, antibiotic-associated diarrhea, and gas gangrene in the livestock producing massive economic losses and a high mortality rate in the animal industry (Azimirad et al., 2019). CPA secretes a metallic enzyme of 43 kDa that exhibits phospholipase C (PLC) and sphingomyelinase (SMase) activities (Oda et al., 2015). In addition, it is a vital immunogenic antigen, which is strongly associated with the pathogenesis of enterotoxaemia and induction of necrotic lesions in the calf intestinal loop model (Goossens et al., 2016). Enterotoxaemia and enterocolitis in sheep and goat and also pulpy kidney diseases in lambs were caused mainly by “ETX” CP-producing strains CP strains of type B and C were also reported in sheep, goat, lambs, and calves with a history of enterotoxaemia and dysentery (Ali Nasir et al., 2015). The biologically active thermolabile trypsin-sensitive toxin produced by CP strains of types “B and C” could be active only in the presence of trypsin inhibitors (Uzal et al., 2018). These toxins were reported in several cases of fatal hemorrhagic dysentery in sheep (type B) and also fatal intestinal necrosis was recorded due to type C infections in in animals and humans (Uzal, 2004). Furthermore, A protoxin of 33 kDa constitutes the ETX (epsilon) toxin of CPD; it is rather inactive when it is created and then be activated with the aid of intestinal protease enzymes to render it fully functional (Navarro et al., 2018). A mature activating form of strong pore-forming toxin that is generated by both B and D types could result in neurologic symptoms, particularly with D strains. Type D was less affiliated with calf enterotoxaemia but it was severely concerned with caprine enterotoxaemia in sheep and goats (Uzal, 2004). Moreover, ETX toxin is a potential biological and intracellular binary toxic warfare agent, which is composed of two distinct proteins (Alves et al., 2014). However, antibiotics could limit successful varieties of bacterial infections, the extensive and indiscriminate use of different classes of antibiotics in the animal field for several purposes led to the spreading of antibiotic-resistant pathogens and so, failure in the treatment strategies (Cao et al., 2020). Natural traditional plants, bacteriophages, prebiotics, and probiotics had attracted attention at the latest recent years as alternative tools for antibiotics (Cao et al., 2020). Moringa. oleifera is one of the best medicinal plants which has been widely used in the treatment of variable sorts of diseases. This plant possesses many sustainable features of naturally derived antibacterial agents. Moringa oleifera is documented with its multiple nutritional, therapeutic, and prophylactic properties (Gopalakrishnan et al., 2016). It is used as a natural substitute for antibiotics with no adverse effects. Moringa oleifera or magic tree or the tree of life (as it is called) is a small to medium-sized tree that has been distributed in many tropical and subtropical countries. Different extracts of their leaves and roots recorded significant reductions in the severity and frequency of diarrhea. It is known for its high antibacterial effect against Gram-positive, and Gram-negative bacteria, spore-forming bacteria, and fungi (Shailemo et al., 2016). Their leaves also acquired effective antibacterial traits against multiple multidrug-resistant (MDR) organisms like Staph aureus, Escherichia coli, and Pseudomonas aeruginosa. Despite many annual reports had discussed the infection rate of CP organisms in different livestock; few published studies were concerned with the broad antibacterial impacts of M. oleifera plant against CP. Hence, this study aimed to investigate the in vitro antibacterial activity of M. oleifera extracts against some virulent CP strains with different toxin types recovered from farm animals (cattle, sheep, goat) in Egypt compared to the commercially available antimicrobial agents already applied in the field. Materials and MethodsSamplingA total of 200 samples were collected in sterile plastic bags from different livestock animals (cattle “n=110,” sheep “n=60,” and goat “n=30”). The samples included: fecal samples from both apparently healthy and diseased animals with diarrhea, liver and intestinal samples from enterotoxemia or freshly dead animals, and soil samples. This cross-sectional survey study was conducted over a 1-year period in different animal farms and sometimes randomly from grazing shepherds or local farmers rearing cattle, sheep, and goats in the Suez Canal area, Egypt. The diseased animals showed signs of illness of enteric diseases with a history of diarrhea) which may include abdominal discomfort, lack of appetite, history of weight loss, and sometimes fever. All the collected samples were transported immediately under refrigerated conditions to the Bacteriological Laboratory at Animal Health Research Institute, Ismailia, Egypt, to be examined. Bacterial scheme for isolation of CPAll samples were diluted in 1:10 phosphate-buffered saline and placed in a water bath for 10 minutes at 80°C to kill the non spore-forming bacteria. The processed samples were sub-cultured in cooked meat broth media (Oxoid, Cambridge, UK) for 24 hours under strict anaerobic conditions for enrichment of CP. After that, these inoculated broth tubes were directly streaked onto sterile freshly prepared blood agar plates (nutrient agar; Oxoid, UK that were supplemented with 5% sterile defibrinated sheep blood and Neomycin sulfate in a concentration of 200 μg/ml). All cultivated blood agar plates were then incubated in anaerobic jars at 37°C for 24 hours using anaerobe sachets (BD GasPak™ EZ Anaerobe Container System Sachets) to produce the strict conditions of anaerobiosis, which must be provided for vitality and viability of CP. Further identification tests for the recovered CP isolates including colony morphology, Gram staining, and biochemical tests (catalase, sugar fermentation, and gelatin liquefaction) were performed. In addition, the lecithinase and lipase activities of these recovered CP isolates on egg yolk agar medium were also examined. In addition, for toxinotyping of the recovered CP isolates, a toxin-antitoxin neutralization test was performed and interpreted according to Quinn et al. (2002). Antimicrobial susceptibility testingAll the recovered CP isolates were in vitro tested for their antimicrobial susceptibility using the agar disc diffusion according to the standards of the Clinical and Laboratory Standards Institute method (CLSI, 2017). Eleven commercial antibiotic disks: Tetracycline (TET; 10 μg), lincomycin (L; 15 μg), norfloxacin (NOR; 10 μg), erythromycin (E; 30 μg), ampicillin (AM; 10 μg), penicillin (P; 10 μg), chloramphenicol (C; 30 μg), amoxicillin clavulanic acid (AMX; 25μg), gentamycin (CEN; 10 μg), cefotaxime (CTX; 30 μg), and ciprofloxacin (CIP; 5 μg); Oxoid, Cambridge, UK; were evaluated and then the results were interpreted according to Gomaa et al. (2023). Molecular toxinotyping of CP isolates with polymerase chain reaction (PCR) techniquePCR was applied to determine the toxin genes (cpa, cpb, and etx), virulence (cpe, Cpb2, tpel, and NetB), and antibiotic resistance genes (TET k, lnu (A), bla, and ermB) of the recovered CP isolates using a specific set of primers as shown in Table 1. The step of the separation of DNA of the recovered isolates was applied first followed by PCR reactions step. It had been performed in the thermal cycler. Table 1 described the primer sequences, product sizes, the accompanied temperatures, and time conditions required for the reaction to amplify the target toxin genes. PCR reactions were calculated in a total volume of 25 ul where 12.5 ul of DreamTaq TM Green Master Mix (2X) (Fermentas, Inc. Hanover, MD, USA), 1ul of each primer [20 pmol (Sigma-Aldrich, Co., St. Louis, MO)], 5 ul of DNA template, and 5.5 ul of PCR grade water were added in the reaction. The final step is gel electrophoresis in which the gel was prepared using 1.5% agarose gel and 10 ul of the tested amplified PCR product of each sample. Then they were inoculated into the gel and then stained with ethidium bromide (0.5 μg/ml) (Sigma-Aldrich, Co., St. Louis, MO) to be easily visualized under UV illumination. Both PCR positive and negative controls were included in each reaction to be easily validated. Sterile saline was considered as a negative control while the positive control was provided by the National Laboratory for Veterinary Quality Control on Poultry Production (NLQP) from previous positive tested and validated field isolates. Table 1. Target antibiotic resistance, toxin, and virulent genes of CP and their primer sequences and expected amplicon sizes.
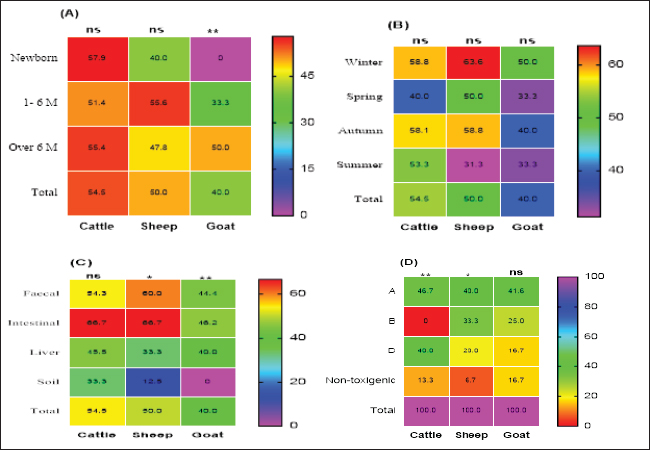
Preparation of M. oleifera plantIn this study, we used Moringa oil and Moringa leaves. The Moringa oil was kindly purchased from CAP-PHARM®, Cairo. While M. oleifera leaves were obtained from sandy soil that was cultivated with Moringa shrubs at Sinai, Egypt. According to Katircioğlu and Mercan Dogan (2006), these leaves were taken under sterile precautions, washed properly, and air-dried. Then, using a sterile mortar and pestle, the dried leaves were grounded, and sieved, and finally, the plant powder was kept in well-tight polyethylene bags until further use. For preparation of the ethanolic and aqueous extracts of Moringa leaves, the previous grounded leaves powder was extracted with 100% ethanol and sterile distilled water. The sample and solvent ratio was 1:2 during extraction. The extracts were collected three times and filtered through Whatman filter paper and then the extracts were kept at 4°C till be needed for the assay of their antibacterial efficacy. Estimation of the antimicrobial efficacy of M. oleifera plantUsing the agar well diffusion method, the antimicrobial activity of previously prepared ethanolic and aqueous extracts of M. oleifera plant was evaluated. The stock cultures 1.5 x 108) of the yielded CP strains in this study were previously prepared separately for each. They were matched with 0.5 Mc-Farland standard then the test was performed. A loopful of each target stock CP strain culture was spread onto sterile separate Muller Hinton agar (MHA) plates and left for 1 hour at 25°C for complete saturation of the organism. Then, using the sterile piercing instrument, make definite holes in the cultivated MHA plates under aseptic conditions so that 50 μl of the prepared ethanolic and aqueous leaf extracts and its oil could be poured, then incubated anaerobically at 37°C for 24 hours. All cultured MHA plates were observed and the diameter of inhibition zones for each were determined, recorded and the results were compared (Abd El-Moez et al., 2014). Statistical analysisThe data analysis was performed using Microsoft Excel (Microsoft Corporation). The prevalence of CP was tested using a chi-square test (Proc freq; SAS Institute Inc., 2012). The software Origin was utilized to generate a cluster dendrogram of different animal types based on the relative frequency of resistance combination to different antibiotics. The differences in the inhibition zones of Moringa oil and extracts against CP isolates in cattle, sheep, and goats were tested according to Mann–Whitney. Furthermore, the U. Roc curve of MedCalc statistical software was used for assessing the AUC values of different diagnostic antibiotics and Cohen’s Kappa test was used to test the quality of agreement between tests. In addition, GraphPad Prism software 9.0 (GraphPad, USA) was used to generate the figures Statistical significance was determined by accepting p-values less than 0.05. Ethical approvalNot needed for this study. ResultsCultural and morphological characteristics of CP isolatesCP isolates displayed turbidity with abundant growth in cooked meat medium broth and gas formation (due to their saccharolytic activity); however, meat particles were pinkish and not digested. On sheep blood agar medium, CP colonies were identified with their characteristic double zone of beta hemolysis (the inner hemolytic clear zone was due to beta toxin while the outer hemolytic one was due to alpha toxin). On egg yolk emulsion agar medium, CP organisms gave opalescence on the side of the plate without antitoxin while it was inhibited on the other side of the plate with antitoxin. Microscopically, they are Gram-positive straight-sided rods arranged singly or in pairs having central or sub-terminal oval nonbulging endospores. In addition, biochemically, CP isolates were catalase and indole negative; however, they were gelatin liquefier and glucose, lactose, and sucrose fermenters. Biochemical results confirmed the identification of CP species. Prevalence and typing of CP in different livestock animalsIn this study, the overall prevalence rate of CP in animals (including cattle, sheep, and goats) was 51% (102/200). CP was found in examined animals with diarrhea or enteritis greater (57.7%) than that in apparently healthy ones (50%) which had not shown any signs of illness. The results of the study revealed significant variations in the prevalence of CP among the examined animals ( p=0.0293) as indicated in Table 2. Moreover, the prevalence rate was declared also in Table 2 and calculated as 57.7% in animals exhibiting diarrhea or enteritis, 54% in animals diagnosed with enterotoxemia or in dead cases, 50% in apparently healthy animals that did not display any signs of illness, and 20% from the surrounding environment (specifically, the soil). Frequency rate of CP in examined animals according to age, host, and seasonal variationCP with variant toxin types were detected in cattle, sheep, and goats in this study of different ages, sampling times, and sample types as depicted in Figure 1. In cattle and sheep, the frequency of CP did not show significant differences across the studied ages. However, in goats, there was a significant difference (p < 0.01), with frequencies of 33.3% and 50% in the age of 1–6 months and over 6 months, respectively, while no cases were detected in newborn goats. Overall, as stated in Figure 1A, animal groups of younger age of all examined animals were more susceptible to CP infections compared to older age. In addition, throughout the year, winter and autumn seasons exhibited higher isolation rates of CP in all examined animal species (Fig. 1B); however, no significant differences were observed between the different seasons studied (p > 0.05). Based on the results of variant types of examined samples in this study, significant differences were found in the prevalence of CP between different samples in sheep and goats (p < 0.05 and 0.01, respectively). The lowest prevalence of CP organisms was observed in soil samples, with 12.5% in sheep; however, it could not be isolated from goat soil samples as shown in Table 2. The highest prevalence was found in intestinal samples, with rates of 46.2% in sheep and 66.7% in goats. In contrast, no significant differences were observed in cattle (p > 0.05) as cleared in Figure 1C. Moreover, regarding toxin typing of CP isolates in cattle, sheep, and goats, type A was the most prevalent at 44.1% (45/102), followed by type D at 31.4% (32/102). Type B was isolated exclusively from sheep and goats, with a frequency of 13/102 at the rate of 12.7%, but it was not detected in cattle as shown in Figure 1D.
Fig. 1. Frequency of CP and their toxin types in cattle, sheep, and goat according to different variants. Table 2. Relation of CP in different animals with animal health conditions.
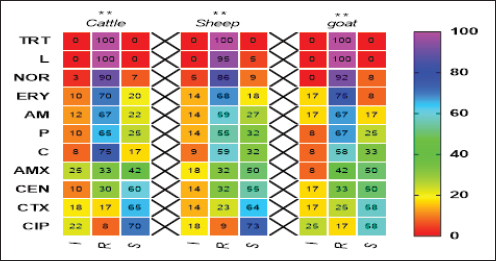
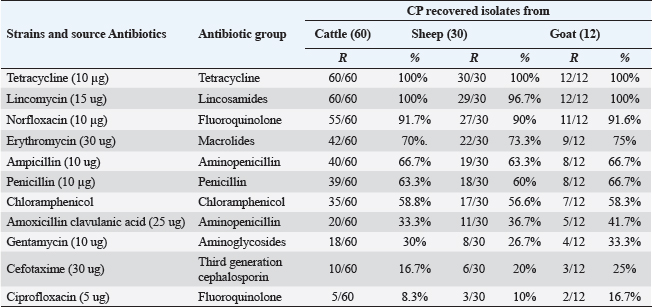
Pos: Positive samples for CP; No.: Number of examined samples. In vitro antimicrobial sensitivity results of the recovered CP isolatesAs shown in Figure 2, the estimated inhibition zones using the disc diffusion agar method classified the tested antibiotics against CP isolates into resistant, intermediate, or sensitive drugs. Accordingly, CP isolates in this study were found to be highly resistant against tetracycline (100%) and lincomycin (96.7%–100%). However, they were of moderate resistance rates against norfloxacin (90%–91.7%), erythromycin (70%–75%), ampicillin (63.3%–66.7%), penicillin (60%–66.7%), chloramphenicol (56.6%–58.8%) and amoxicillin-clavulanic acid (33.3%–41.7%) in all animal species as estimated in Table 3. In addition, ciprofloxacin, cefotaxime, and gentamycin drugs were the most sensitive drugs showing the lowest resistance rate that ranged (from 8.3% to 16.7%), (from 16.7% to 25%), and (from 26.7% to 33.3%) against CP isolates in all examined animals as estimated in Table 3.
Fig. 2. Antimicrobial resistance pattern of different antibiotics against different CP isolates. (I, intermediate; R, resistance, S, sensitive); **p < 0.01. (TET): Tetracycline; (L): lincomycin; (NOR): norfloxacin; (E): erythromycin; (AM): ampicillin; (P): penicillin; (C): chloramphenicol; (AMX): amoxicillin clavulanic acid; (CEN): gentamycin; (CTX): cefotaxime; (CIP): ciprofloxacin. Table 3. Resistance patterns of CP isolates for most commercial antibiotics.
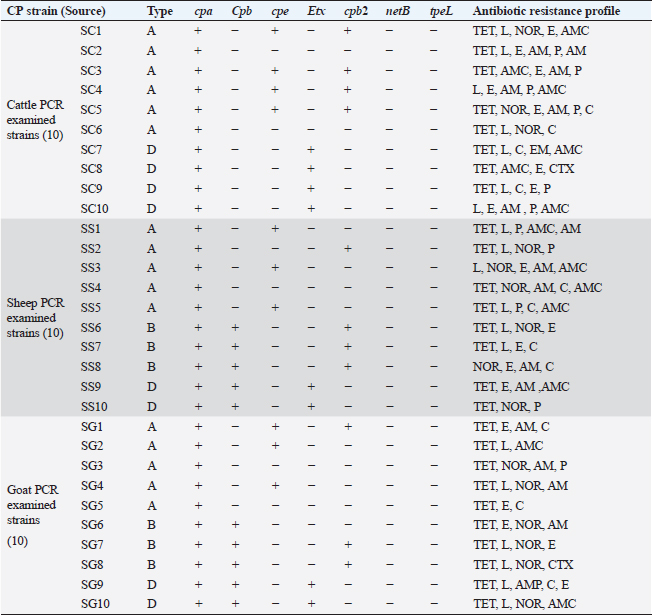
PCR toxin genotyping and resistance determinants of CP isolatesThe distribution of different toxin, virulent, and antibiotic-resistant genotypes of the 10 PCR-selected CP strains in this study compared with their phenotype and their antibiotic resistance pattern was depicted in Table 4. PCR confirmed that all selected isolates of CP were of type A as they all carried cpa gene; however, cpb gene was detected in (type B only) in sheep and goats and type D exhibited their genes of Cpa, Cpb, and Etx toxin genes altogether in some isolates. For virulence determinants, the enterotoxin (Cpe) gene was found in 40%, 30%, and 30% of tested ten strains in cattle, sheep, and goat species, respectively. Meanwhile, the beta2 toxin (Cpb2) gene was discovered in 40% of each of the tested cattle and sheep CP isolates but in 30% of ten tested goat strains. Interestingly, PCR could not detect netB and tpeL virulent genes at all in the tested strains in all species. Table 4. Distribution of toxin genotyping, virulence determinants, and antibiotic resistance profiles in different isolates of CP.
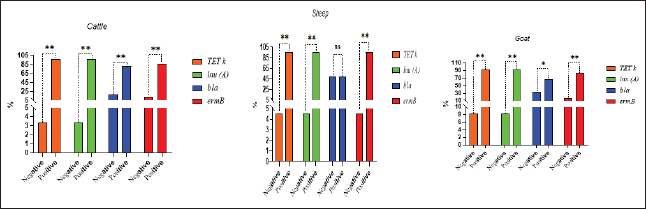
SC: CP strains from cattle; SS: CP strains from sheep; SG: CP strains from goat; TET: Tetracycline; L: lincomycin; NOR: norfloxacin; E: erythromycin; AM: ampicillin; P: penicillin; C: chloramphenicol; AMX: amoxicillin clavulanic acid; CEN: gentamycin; CTX: cefotaxime; CIP: ciprofloxacin. Analysis of resistant genes of CP isolates and their toxin typesA sequential arrangement of the different types of CP toxin with some antibiotic-resistant genes was clarified in Figure 3. The lincomycin and tetracycline-resistant [lnu (A) and TET k] genes were closely clustered together, followed by erythromycin-resistant (ermB) gene. However, bla gene (β-lactam resistant) occupied the third position, forming a cluster with the three genes. Statistically significant differences were observed between negative and positive cases for toxin types and virulence determinants (p < 0.05) except bla in sheep showed nonsignificant differences (p > 0.05).
Fig. 3. Analysis of different toxin types of CP isolates and resistance determinants in different species; ns, nonsignificant; *p < 0.05; **p < 0.01.
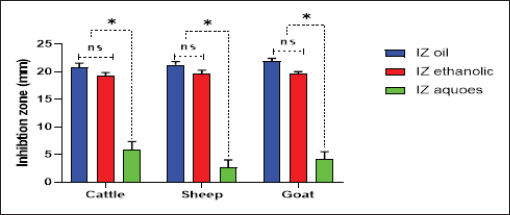
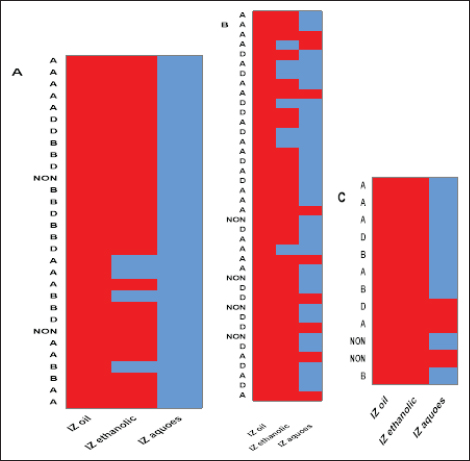
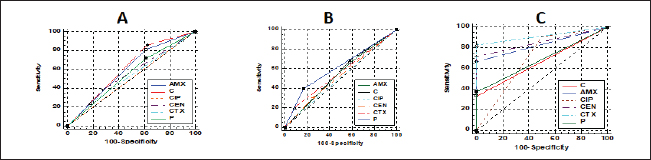
Fig. 4. Antibacterial efficacy of M. oleifera oil and extracts according to their inhibition zones (IZ) against CP isolates in cattle sheep and goats. In vitro evaluation of the antimicrobial activities of Moringa plantThe antibacterial efficacy of M. oleifera oil and extracts against CP isolates from different animals was studied. According to agar well diffusion results, the oil and ethanolic extracts of M. oleifera leaves were of high antibacterial potency as shown in Figure 4. The oil and the ethanolic treatment of M. oleifera produced significantly higher zones of inhibition that ranged from 20 to 24 mm than the aqueous extracts (which did not exceed 10 mm) against CP in cattle, sheep, and goat (p < 0.05). Nonsignificant differences were observed between the M. oleifera oil and the ethanolic extract in all studied strains. Correlation of M. oleifera oil and extracts and toxin types of CP isolatesBased on the animal species and toxin type of CP, Figure 5 showed that the antibacterial efficacy of M. oleifera oil had been strongly correlated with all types of the recovered isolates particularly type A in (cattle, sheep, and goat) species. However, ethanol and aqueous extracts of the plant leaves correlated in lesser values. Correlation between sensitivity results of M. oleifera oil and extracts and most tested antibiotics against CP isolatesThe relation between the sensitivity results by aqueous, ethanolic extract and oil of M. oleifera against CP isolates in three tested animal species (cattle, sheep, and goat) was demonstrated in Figure 6A–C, respectively. The analysis of the area under this curve in Figure 6A discussed similar efficacy of the aqueous extract of M. oleifera with both chloramphenicol and amoxicillin clavulanic acid drugs which were closely correlated with it. However, the sensitivity of the cefotaxime drug was closely correlated with that produced by the ethanolic extract of M. oleifera plant against CP isolates as shown in Figure 6B. Moreover, ciprofloxacin and gentamycin drugs emerged highest sensitivity that was equal to that given by the oil of M. oleifera (Fig. 6C).
Fig. 5. Relationships between different treatments of Moringa plant (M. oleifera) and toxins of CP isolates from sheep (a), cattle (b), and goat (c). Red cells indicate the sensitive antibacterial effects of different Moringa treatments (oil or extracts) while blue cells indicate the resistance pattern of Moringa oil or other extracts. (A) Indicates CP isolates of type A, (B) indicates CP isolates of type B, (D) indicates CP isolates of type D, (NON) indicates nontoxigenic strains. IZ oil: inhibition zones that were recorded by the oil of the Moringa plant. IZ ethanolic: inhibition zones that were recorded by ethanolic extract of the Moringa plant. IZ aqueous: inhibition zones that were recorded by aqueous extract of the Moringa plant.
Fig. 6. Relationships between different treatments of Moringa (Aqueous; A, Ethanol; B, and oil; C) and antibiotics (Penicillin (P), Chloramphenicol (C), Amoxicillin clavulanic acid (AMX), Gentamycin (CEN), Cefotaxime (CTX), and Ciprofloxacin (CIP) for CP isolates. DiscussionCP are serious pathogens intended for many animal histotoxic and enteric diseases (Fohler et al., 2016). The results of the current investigation revealed that CP was isolated in 51% of the total 200 examined samples from cattle, sheep, and goat species with the highest prevalence rate of CP strains from cattle species (54.5%) more than in sheep (50%) and goat (40%). Similarly, CP was isolated from 90/200 to 70/200 cattle and sheep samples were also documented in southern Iran (Hosseinzadeh et al., 2018). It was isolated also by 49% from the total of 174/355 rectal swabs of examined cattle, sheep, and goat animals. Moreover Kronfeld et al. (2022) detected CP pathogens in 14/27 (51.9%) in cow cases with a pathological puerperium. On the other hand, the higher incidence of CP 70.62% was recorded in 125/177 of the fecal samples of healthy, diarrheic, and morbid animals in Kashmir valley in India; of which, 110/125 (72.36%) were sheep isolates while 15 (60%) were goat isolates (Nazki et al., 2017). The discrimination in the incidence rate might be due to different geographic distribution or environmental climatic change, bad sanitary or hygienic conditions, lack of rapid diagnostic tests, or might be due to variant duration of the investigations studies (Moustafa et al., 2022). In Egypt, CP was reported in 51.5% where 103 out of a total of 200 examined lambs (100 lambs with diarrhea, 60 freshly dead, and 40 apparent healthy) showed the organisms (Moustafa et al., 2022). The recorded percentage (60%) in this study among diseased lambs came in line with that of 69.29% in a study of Kumar et al. (2014). In addition, in Pakistan, CP was identified in healthy and diseased sheep and goats in 46.1% (Mohiuddin et al., 2020). According to the health status of examined animals, the prevalence rate of CP in the current study was stated in Table 2 in the apparently healthy cattle (46.7%), sheep (60%), and goats (33.3%). This was nearly like the finding of Hamza et al. (2018) who found CP pathogens in the rectal swabs of apparently healthy sheep, buffaloes, and cattle in percentages of 65.45%, 55%, and 47.1%, respectively, at Cairo and Giza governorates in Egypt. CP is one of the heterogeneous groups of environmental bacteria that inhabitant in the animal surroundings, its environment, soil, and water since its existence might indicate the animal fecal contamination (Omar et al., 2018). The clostridia spores in soils and/or in apparently healthy animals could make sporadic disease episodes that could result in massive economic losses in animal production via the ingestion route and then production of their toxins (Gamboa et al., 2005; Diego et al., 2012). Great attention should be paid to the role of soil and or other surrounding factors like the irrigated water posing a risk especially when the water and soil contact food in the field. CP bacterium was isolated from the soil and irrigated water in 40% and 31.7% of samples, respectively (Hamza et al., 2018). This pathogen could affect all ages of animals (Nazki et al., 2017) but younger ages were severely affected (Jemal et al., 2016). Figure 1A cleared that the highest incidence of infection was at young ages in all examined species and this was consistent with Omar et al. (2018) who stated that newly born lambs with the age of <3 months were more susceptible to clostridial infections than older lambs (3–6 months and over 6 months of age). Regarding the seasonal variation and their effects on the incidence of CP infections in animals, the recent results in Figure 1B showed a higher incidence rate in winter (58.2%) followed by autumn season (56.7%). This corresponds to many studies in sheep and calves, respectively (Selim et al., 2017; Moustafa et al., 2022), in which, they confirmed that the winter season was of the highest incidence all over the year. This finding might be attributed to poor hygienic conditions, lower temperatures, and changes in the pasture during the winter season in comparison with other seasons (Omar et al., 2018). Enterotoxaemia is an acute, highly fatal disease that could produce severe losses in animal production. It affects all ages of animals (Nazki et al., 2017). In this study, the prevalence rate of CP was estimated in Figure 1C; from a total of 80 fecal and 54 intestinal and liver samples. A recent study in Saudi Arabia found that the prevalence of enterotoxemia was lowest in sheep (21.4%) compared with cattle, goats, and camels. These results were the same way with Alsaab et al. (2021) who detected it in 43% (40/93) from total examined 93 rectal swabs and intestinal content samples in diseased and enterotoxemic animals. Moreover, a high mortality rate in sheep and goats with enterotoxaemia in Egypt at El-Behera Governorate was referred to CPA organisms with its alpha toxin from 104 intestinal, liver, kidney, and spleen of the suspected cases. In addition, compatible results were reported by Nazki et al. (2017) and Omer et al. (2020). Application of PCR in toxin-genotyping of CP could provide a real basis for rapid diagnosis of this bacteria in animal farming (Ibrahim et al., 2017; Milanov et al., 2018). In this study, PCR confirmed the toxinotyping of the recovered CP in cattle, sheep, and goat strains. PCR displayed that type A was the most prevalent toxin in all examined animals followed by D and B as shown in Figure 1D. Many reports confirmed that finding of type A of CP was the highly detected type in cattle and sheep samples (Elsify et al., 2016; Hosseinzadeh et al., 2018; Hayati et al., 2020) . Moreover, Nazki et al. (2017) detected also that most of the sheep and goat isolates were type A (60.90% and 53.33%) followed by type D (39.09% and 46.66%), respectively. In the same trend, the typing of 75 recovered CP isolates of multi-species indicated that type A was the major identified type (90.67%) (Anju et al., 2021). In addition, Omer et al. (2020) confirmed that CP isolates were identified in 80.8% of type A, 15.4% of D, 2.9% of type C, and 0.98% only for type B. The exponential long-term use of antibiotics in the animal field has played the main role in the rapid emergence of resistance traits among CP isolates. That resistance might be conferred by various genes transferring the resistance to other bacteria within the genus or to different genera (Anju et al., 2021). According to the estimated diameter of the inhibition zones of CP isolates in Figure 2 and Table 3, CP isolates were found to mostly be resistant to multiple antimicrobials in varying degrees in all species. The highest resistance ranged between 96.7%–100% was recorded for tetracycline and lincomycin; however, the isolates showed the highest sensitivity to ciprofloxacin, cefotaxime, and gentamycin drugs. Moreover, the phenotypic and genotypic of some resistant antibiotic correlations for CP isolates in this study were discussed in detail in Table 4, and also, the correlation between resistant genes and different types of isolates was also reported in Figure 3. Corresponding antimicrobial sensitivity results of CP isolates reported in Anju et al. (2021) in which CP showed high resistance towards multiple antimicrobials. They were resistant to gentamicin (44%), erythromycin (40%), and tetracycline (26.67%). Similar high-resistance tetracycline was reported by Yadav et al. (2017) and Gharieb et al. (2021). Another study by Mohiuddin et al. (2020) clarified that all 184 yielded CP isolates in small ruminants in Pakistan were 100% resistant to neomycin and 72% resistance to tetracycline mean while ciprofloxacin and erythromycin drugs showed least sensitivity rates (43% and 14%) and 57% showed sensitivity to teicoplanin, chloramphenicol, amoxicillin, linezolid, and enrofloxacin; however, 100% of the isolates were sensitive to rifampin and ceftiofur. They declared that a higher incidence of antibiotic resistance in CP isolates might result from the excessive use of this antibiotic in the sampling areas, often following poor or incorrect veterinary advice. Moringa plant (M. oleifera) is a promising biomolecule. It could act as a potential drug candidate that might suppress several bacteria species including pathogenic Clostridium spp. and also succeed in the treatment of gastroenteritis, diarrhea, and other disorders (Adji et al., 2022). Moringa oleifera plant extract achieved excellent antimicrobial results against Clostridium spp. in the diseased sheep and significantly lowered the fecal bacterial count of C. novyi in the examined cases (El Shanawany et al., 2019). As mentioned in Figure 4, the corresponding results of M. oleifera plant in this study provided potent evidence that enhanced its use for medicinal purposes as a strong antibacterial drug against CP. The potent antibacterial role of this plant to the presence of an array of phytochemicals in their leaves identifying a short peptide 4 (´a -L-rhamnosyloxy) benzyl-isothiocyanate compounds that might play the main role in the inhibition of the microbial growth through disruption of the synthesis of the cell membrane or impaired important enzymes. Furthermore, the type of solvent in the extraction method could impact the antimicrobial potency of the M. oleifera (Fig. 4). Hence, in the current study, the ethanolic extract produced better antimicrobial results than the aqueous one. In the same way, the extracts of the leaves and seeds of the M. oleifera against pathogenic Clostridium spp. inhibited their growth in varying degrees depending on the solvent employed in extraction. The aqueous extract was of lesser antibacterial activity against Clostridium spp. than ethanolic and acetone extracts of the fresh green stemmed leaf that were more active at lower concentrations. The decoction of the plant parts in the water might not be an effective method if compared with the plant preparation method via an organic material as a solvent which this type of solvent could strengthen the antibacterial activity of this plant (Abd El-Moez et al., 2014). Adji et al. (2022) explained the great antibacterial properties that plants containing high natural components: (pterygospermin, benzyl isothiocyanate, and 4 L-rhamnopyra nosyloxy and benzylglucosinolate) that possessed high antibacterial activity. In addition, the antibacterial effect of M. oleifera oil was highly proportional with CP of type A mainly than other types in all tested species than ethanol and aqueous extracts that were of lesser effect as shown in Figure 5. Moreover, the sensitivity results of different treatments of the Moringa plant (oil, ethanolic, and aqueous extracts) compared to that of tested antibiotics against the recovered CP based on agar well diffusion method were reported in Figure 6 and indicated similar high efficacy of M. oleifera oil and ethanolic extracts as potent antibacterial against CP isolates. Broad-spectrum antibacterial efficacy of the M. oleifera plant was confirmed also against most pathogenic Gram-positive and Gram-negative organisms in many reports in which greater inhibition zones of Moringa were recorded against S. aureus and E. coli isolates (Abd El-Moez et al., 2014). In addition, recent reviews declared that the ethanol and methanol extracts of M. oleifera could suppress the viability of Salmonella typhi, Salmonella paratyphi, E. coli, B. cereus, Shigella, S. aureus, and E. faecalis organisms (Híjar et al., 2018; Salihu Abdallah et al., 2019; Abdallah et al., 2022; Adji et al., 2022) than the aqueous extract. The active components responsible for the bactericidal activity are more soluble in organic solvents than water extract (Prabakaran et al., 2018). Moreover, a significant minimum inhibitory concentration (MIC) of Ethanolic leaves’ extract (79%–0.3%) was detected against P. aeruginosa which could be owed to the high total phenolic content that might interact with the protein and enzymes of the cell membrane destroying the cell membranes structures and inhibiting its functions causing the microbial death (Mostafa et al., 2018). ConclusionThe current findings in this study deliberated a higher prevalence of MDR and more pathogenic CP strains from different livestock. Therefore, epidemiological investigations, sanitary measures, prophylaxis plans, and control strategies in animal Egyptian farms should be adequately enforced. Moreover, M. oleifera had come into the limelight as it was a potent antibacterial herbal medicinal plant and offered the best antimicrobial results in our study against CP indicating that it could be a better antibiotic substitute in animal husbandry. More detailed studies for the pharmacokinetics of M. oleifera plant were recommended also for evaluating its explicit role in limiting CP toxicity and pathogenicity in animals. AcknowledgmentThe authors are thankful to Prof. Dr. Momtaz Shahin, the president of the Animal Health Research Institute (AHRI), Agriculture Research Center (ARC), Egypt, Dokii, Giza. Also, we are thankful to all our colleagues in AHRI, Ismailia branch for their encouragement and assistance. Conflict of interestThere is no conflict of interest. FundingNone. Authors’ contributionsGAI and KAA: Conceived and designed the study. GAI and KAA: Performed the study. GAI and KAA: Wrote the manuscript, Analyzed the data, Drafted, updated the references, and revised the manuscript. All authors have read and approved the manuscript. Data availabilityAll data are provided in the manuscript. ReferencesAbd El-Moez, S., Badawy, A.Y.E. and Omer, H. 2014. Assessment of antimicrobial effect of Moringa: In vitro and in vivo evaluation. African. J. Microbiol. Res. 8, 3630–3638. Abdallah, M., Machina, F.M. and Ibrahim S. 2022. Antibacterial activity of Moringa oleifera methanolic leaves extracts against some Gram-positive and Gram-negative bacterial isolates. Microb. Infect. Dis. 3, 199–208. Adji, A.S., Atika, N., Kusbijantoro, Y.B., Billah, A., Putri, A. and Handajani, F. 2022. A review of leaves and seeds Moringa oleifera extract: the potential Moringa oleifera as antibacterial, anti-inflammatory, antidiarrhoeal, and antiulcer approaches to bacterial gastroenteritis. Open Access Macedonian J. Med. Sci. 10, 305–313. Ali Nasir, A., Younus, M., Rashid, A., Abdul Khaliq, S., Khan, E., Shah, S.H., Aslam, A., Ghumman, M.A. and Joiya, M.H. 2015. Clinico-pathological findings of Clostridium perfringens type D enterotoxaemia in goats and its hemolytic activity in different erythrocytes. Iran. J. Vet. Res. 16, 94–99. Alsaab, F., Wahdan, A. and Saeed, E.M.A. 2021. Phenotypic detection and genotyping of Clostridium perfringens associated with enterotoxemia in sheep in the Qassim Region of Saudi Arabia. Vet. World. 14, 578–584. Alves, G.G., Machado de Ávila, R.A., Chávez-Olórtegui, C.D. and Lobato, F.C.F. 2014. Clostridium perfringens epsilon toxin: the third most potent bacterial toxin known. Anaerobe 30, 102–107. Aminov, R.I., Garrigues-Jeanjean, N. and Mackie, R.I. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67, 22–32. Anju, K., Karthik, K., Divya, V., Mala Priyadharshini, M.L., Sharma, R.K. and Manoharan, S. 2021. Toxinotyping and molecular characterization of antimicrobial resistance in Clostridium perfringens isolated from different sources of livestock and poultry. Anaerobe 67, 102298. Azimirad, M., Gholami, F., Yadegar, A., Knight, D.R., Shamloei, S., Aghdaei, H.A. and Zali, M.R. 2019. Prevalence and characterization of Clostridium perfringens toxinotypes among patients with antibiotic-associated diarrhea in Iran. Sci. Rep. 9, 7792. Bailey, M.A., Macklin, K.S. and Krehling, J.T. 2013. Use of a multiplex PCR for the detection of toxin-encoding Ggnes netB and tpeL in strains of Clostridium perfringens. ISRN. Vet. Sci. 2013, 865702. Bueschel, D.M., Helen Jost, B., Billington, S.J., Trinh, H.T. and Glenn Songer, J. 2003. Prevalence of cpb2, encoding beta2 toxin, in Clostridium perfringens field isolates: correlation of genotype with phenotype. Vet. Microbiol. 94, 121–129. Cao, X., Tang, L., Zeng, Z., Wang, B., Zhou, Y., Wang, Q., Zou, P. and Li, W. 2020. Effects of probiotics BaSC06 on intestinal digestion and absorption, antioxidant capacity, microbiota composition, and macrophage polarization in pigs for fattening. Front. Vet. Sci. 7, 570593. Catalán, A., Espoz, M.C., Cortés, W., Sagua, H., González, J. and Araya, J.E. 2010. Tetracycline and penicillin resistant Clostridium perfringens isolated from the fangs and venom glands of Loxosceles laeta: its implications in loxoscelism treatment. Toxicon 56, 890–896. CLSI. 2017. Performance standards for antimicrobial susceptibility testing. Twenty-Seventh Informational Supplement; CLSI Document M100-S27. Wayne, PA: CLSI. El Shanawany, E.E., Fouad, E.A., Keshta, H.G., Hassan, S.E., Hegazi, A.G. and Abdel-Rahman, E.H. 2019. Immunomodulatory effects of Moringa oleifera leaves aqueous extract in sheep naturally co-infected with Fasciola gigantica and Clostridium novyi. J. Parasit. Dis. 43, 583–591. Elsify, A., Tarabees, R., Nayel, M., Salama, A., Aly, M., Mohamed, M., Hassan, H., Zaghawa, A. and Elballal, S. 2016. Bacteriological and molecular studies on Clostridium perfringens isolated from sheep in three Egyptian provinces. African. J. Microbiol. Res. 10, 725–732. Fohler, S., Klein, G., Hoedemaker, M., Scheu, T., Seyboldt, C., Campe, A., Jensen, K.C. and Abdulmawjood, A. 2016. Diversity of Clostridium perfringens toxin-genotypes from dairy farms. BMC. Microbiol. 16, 199. Gharieb, R., Saad, M., Abdallah, K., Khedr, M., Farag, E. and Abd El-Fattah, A. 2021. Insights on toxin genotyping, virulence, antibiogram profiling, biofilm formation and efficacy of disinfectants on biofilms of Clostridium perfringens isolated from poultry, animals and humans. J. Appl. Microbiol. 130, 819–831. Gomaa, N.H., El-Aziz, N.K.A., El-Naenaeey, E.-s.Y., Abdelaziz, W.S. and Sewid, A.H. 2023. Antimicrobial potential of myricetin-coated zinc oxide nanocomposite against drug-resistant Clostridium perfringens. BMC. Microbiol. 23, 79. Goossens, E., Verherstraeten, S., Valgaeren, B.R., Pardon, B., Timbermont, L., Schauvliege, S., Rodrigo-Mocholí, D., Haesebrouck, F., Ducatelle, R., Deprez, P.R. and Van Immerseel, F. 2016. The C-terminal domain of Clostridium perfringens alpha toxin as a vaccine candidate against bovine necrohemorrhagic enteritis. Vet. Res. 47, 52. Gopalakrishnan, L., Doriya, K. and Kumar, D.S. 2016. Moringa oleifera: A review on nutritive importance and its medicinal application. Food. Sci. Human. Wellness. 5, 49–56. Hamza, D., Dorgham, S.M., Elhariri, M., Elhelw, R. and Ismael, E. 2018. New insight of apparently healthy animals as a potential reservoir for Clostridium Perfringens: a public health implication. J. Vet. Res. 62, 457–462. Hayati, M., Shamseddini, M., Tahamtan, Y., Sadeghzadeh, S., Manavian, M. and Nikoo, D. 2020. Isolation and toxin typing of Clostridium Perfringens from sheep, goats, and cattle in fars Province, Iran. Int. J. Enteric. Pathog. 8, 89–93. Hosseinzadeh, S., Bahadori, M., Montaseri, M., Dehghani, M., Fazeli, M. and Nazifi, S. 2018. Molecular characterization of Clostridium perfringens isolated from cattle and sheep carcasses and its antibiotic resistance patterns in Shiraz Slaughterhouse, Southern Iran. Vet. Arhiv. 88, 581–591. Ibrahim, G.A., Shalaby, B., Ammar, A. and Youssef, F. 2017. Toxin genotyping of C. perfringens isolated from broiler cases of necrotic enteritis. Anim. Vet. Sci. 5, 108-120. Jemal, D., Shifa, M. and Kebede, B. 2016. Review on pulpy kidney disease. J. Vet. Sci. Technol. 7, 1–6. Katircioğlu, H. and Mercan Dogan, N. 2006. Antimicrobial activity and chemical compositions of Turkish propolis from different region. Afr. J. Biotechnol. 5, 1151–1153. Kronfeld, H., Kemper, N. and Hölzel, C.S. 2022. Phenotypic and genotypic characterization of C. perfringens isolates from dairy cows with a pathological puerperium. Vet. Sci. 9, 173. Kumar, N., Sreenivasulu, D. and Narsimha reddy, Y. 2014. Prevalence of Clostridium perfringens toxin genotypes in enterotoxemia suspected sheep flocks of Andhra Pradesh. Vet. World. 7(12), 1132–1136. Masco, L., Van Hoorde, K., De Brandt, E., Swings, J. and Huys, G. 2006. Antimicrobial susceptibility of Bifidobacterium strains from humans, animals and probiotic products. J. Antimicrob. Chemother. 58, 85–94. Meer, R.R. and Songer, J.G. 1997. Multiplex polymerase chain reaction assay for genotyping Clostridium perfringens. Am. J. Vet. Res. 58, 702–705. Milanov, D., Petrovic, T., Todorović, D., Aleksic, N. and Cabarkapa, I. 2018. Toxin genotypes of Clostridium perfringens in animal feed and their role in the ethiology of enterotoxemia in domestic animals. Food. Feed. Res. 45, 67–76. Milton, A.A.P., Agarwal, R.K., Bhuvana Priya, G., Saminathan, M., Aravind, M., Reddy, A., Athira, C.K., Ramees, T., Sharma, A.K. and Kumar, A. 2017. Prevalence and molecular typing of Clostridium perfringens in captive wildlife in India. Anaerobe 44, 55–57. Mohiuddin, M., Iqbal, Z., Siddique, A., Liao, S., Salamat, M.K.F., Qi, N., Din, A.M. and Sun, M. 2020. Prevalence, genotypic and phenotypic characterization and antibiotic resistance profile of Clostridium perfringens type A and D isolated from feces of sheep (Ovis aries) and Goats (Capra hircus) in Punjab, Pakistan. Toxins 12, 657. Mostafa, A.A., Al-Askar, A.A., Almaary, K.S., Dawoud, T.M., Sholkamy, E.N. and Bakri, M.M. 2018. Antimicrobial activity of some plant extracts against bacterial strains causing food poisoning diseases. Saudi. J. Biol. Sci. 25, 361–366. Moustafa, S., Zakaria, I., Moustafa, A., AboSakaya, R. and Selim, A. 2022. Bacteriological and serological investigation of Clostridium perfringens in lambs. Sci. Rep. 12, 19715. Navarro, M.A., McClane, B.A. and Uzal, F.A. 2018. Mechanisms of action and cell death associated with Clostridium perfringens Toxins. Toxins 10, 212. Nazki, S., Wani, S.A., Parveen, R., Ahangar, S.A., Kashoo, Z.A., Hamid, S., Dar, Z.A., Dar, T.A. and Dar, P.A. 2017. Isolation, molecular characterization and prevalence of Clostridium perfringens in sheep and goats of Kashmir Himalayas, India. Vet. World. 10, 1501–1507. Oda, M., Terao, Y., Sakurai, J. and Nagahama, M. 2015. Membrane-binding mechanism of Clostridium perfringens Alpha-Toxin. Toxins 7, 5268–5275. Ohtani, K. and Shimizu, T. 2016. Regulation of Toxin Production in Clostridium perfringens. Toxins (Basel) 8, 207. Omar, A.A., Baker, N.M., Bkheet, A.A., Khder, A.M. and Nasr, M.Y. 2018. Epidemiological studies and molecular characterization of Clostridium perfringens in small ruminant at El-Behera governorate, Egypt. Assiut. Vet. Med. J. 64, 81–88. Omer, S.A., Al-Olayan, E.M., Babiker, S.E.H., Aljulaifi, M.Z., Alagaili, A.N. and Mohammed, O.B. 2020. Genotyping of Clostridium perfringens isolates from domestic livestock in Saudi Arabia. Bio. Med. Res. Int. 2020, 9035341. Prabakaran, M., Kim, S.-H., Sasireka, A., Chandrasekaran, M. and Chung, I.-M. 2018. Polyphenol composition and antimicrobial activity of various solvent extracts from different plant parts of Moringa oleifera. Food. Biosci. 26, 23–29. Quinn, P., Markey, B.K., Carter, M., Donnelly, W. and Leonard, F. 2002. Veterinary microbiology and microbial disease. Blackwell science. Can. Vet. J. 44(12), 986. Rocchetti, G., Pagnossa, J.P., Blasi, F., Cossignani, L., Hilsdorf Piccoli, R., Zengin, G., Montesano, D., Cocconcelli, P.S. and Lucini, L. 2020. Phenolic profiling and in vitro bioactivity of Moringa oleifera leaves as affected by different extraction solvents. Food. Res. Int. 127, 108712. Salihu Abdallah, M., Ali, M. and Ms, A. 2019. Antibacterial activity of Moringa oleifera leaf extracts against bacteria isolated from patients attending general sani abacha specialist hospital Damaturu. All. Pharm. Sci. 1, 61–66. Selim, A.M., Elhaig, M.M., Zakaria, I. and Ali, A. 2017. Bacteriological and molecular studies of Clostridium perfringens infections in newly born calves. Trop. Anim. Health. Prod. 49, 201–205. Shailemo, D.H.P., Kwaambwa, H.M., Kandawa-Schulz, M. and Msagati, T.A.M. 2016. Antibacterial activity of Moringa ovalifolia and Moringa oleifera methanol, N-hexane and water seeds and bark extracts against pathogens that are implicated in water borne diseases. Green. Sustainable. Chem. 06, 71–77. Soge, O.O., Tivoli, L.D., Meschke, J.S. and Roberts, M.C. 2009. A conjugative macrolide resistance gene, mef(A), in environmental Clostridium perfringens carrying multiple macrolide and/or tetracycline resistance genes. J. Appl. Microbiol. 106, 34–40. Uzal, F.A. 2004. Diagnosis of Clostridium perfringens intestinal infections in sheep and goats. Anaerobe 10, 135–143. Uzal, F.A., Navarro, M.A., Li, J., Freedman, J.C., Shrestha, A. and McClane, B.A. 2018. Comparative pathogenesis of enteric clostridial infections in humans and animals. Anaerobe 53, 11–20. Yadav, J.P., Das, S.C., Dhaka, P., Vijay, D., Kumar, M., Mukhopadhyay, A.K., Chowdhury, G., Chauhan, P., Singh, R., Dhama, K., Malik, S.V.S. and Kumar, A. 2017. Molecular characterization and antimicrobial resistance profile of Clostridium perfringens type A isolates from humans, animals, fish and their environment. Anaerobe 47, 120–124. | ||
| How to Cite this Article |
| Pubmed Style Ibrahim GA, Altammar KA, . Moringa oleifera as a potential antimicrobial against pathogenic Clostridium perfringens isolates in farm animals. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 242-255. doi:10.5455/OVJ.2024.v14.i1.21 Web Style Ibrahim GA, Altammar KA, . Moringa oleifera as a potential antimicrobial against pathogenic Clostridium perfringens isolates in farm animals. https://www.openveterinaryjournal.com/?mno=176977 [Access: May 08, 2024]. doi:10.5455/OVJ.2024.v14.i1.21 AMA (American Medical Association) Style Ibrahim GA, Altammar KA, . Moringa oleifera as a potential antimicrobial against pathogenic Clostridium perfringens isolates in farm animals. Open Vet J. 2024; 14((1) (Zagazig Veterinary Conference)): 242-255. doi:10.5455/OVJ.2024.v14.i1.21 Vancouver/ICMJE Style Ibrahim GA, Altammar KA, . Moringa oleifera as a potential antimicrobial against pathogenic Clostridium perfringens isolates in farm animals. Open Vet J. (2024), [cited May 08, 2024]; 14((1) (Zagazig Veterinary Conference)): 242-255. doi:10.5455/OVJ.2024.v14.i1.21 Harvard Style Ibrahim, G. A., Altammar, K. A. & (2024) Moringa oleifera as a potential antimicrobial against pathogenic Clostridium perfringens isolates in farm animals. Open Vet J, 14 ((1) (Zagazig Veterinary Conference)), 242-255. doi:10.5455/OVJ.2024.v14.i1.21 Turabian Style Ibrahim, Ghada A., Khadijah A. Altammar, and . 2024. Moringa oleifera as a potential antimicrobial against pathogenic Clostridium perfringens isolates in farm animals. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 242-255. doi:10.5455/OVJ.2024.v14.i1.21 Chicago Style Ibrahim, Ghada A., Khadijah A. Altammar, and . "Moringa oleifera as a potential antimicrobial against pathogenic Clostridium perfringens isolates in farm animals." Open Veterinary Journal 14 (2024), 242-255. doi:10.5455/OVJ.2024.v14.i1.21 MLA (The Modern Language Association) Style Ibrahim, Ghada A., Khadijah A. Altammar, and . "Moringa oleifera as a potential antimicrobial against pathogenic Clostridium perfringens isolates in farm animals." Open Veterinary Journal 14.(1) (Zagazig Veterinary Conference) (2024), 242-255. Print. doi:10.5455/OVJ.2024.v14.i1.21 APA (American Psychological Association) Style Ibrahim, G. A., Altammar, K. A. & (2024) Moringa oleifera as a potential antimicrobial against pathogenic Clostridium perfringens isolates in farm animals. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 242-255. doi:10.5455/OVJ.2024.v14.i1.21 |