| Original Article Online Published: 15 Aug 2020 | ||
Open Vet. J.. 2020; 10(3): 276-288 doi: 10.4314/ovj.v10i3.6 Open Veterinary Journal, (2020), Vol. 10(3): 276–288 Original Research http://dx.doi.org/10.4314/ovj.v10i3.6 Extent of pathogenic and spoilage microorganisms in whole muscle meat, meat products and seafood sold in Libyan marketHanan L. Eshamah1, Hesham T. Naas1, Aboubaker M. Garbaj1, Salah M. Azwai2, Fatim T. Gammoudi2, Ilaria Barbieri3 and Ibrahim M. Eldaghayes2*1Department of Food Hygiene and Control, Faculty of Veterinary Medicine, University of Tripoli, Tripoli, Libya 2Department of Microbiology and Parasitology, Faculty of Veterinary Medicine, University of Tripoli, Tripoli, Libya 3Department of Genetics, The Lombardy and Emilia Romagna Experimental Zootechnic Institute, Brescia, Italy *Corresponding Author: Ibrahim Eldaghayes. Department of Microbiology and Parasitology, Faculty of Veterinary Medicine, University of Tripoli, Tripoli, Libya. Email: ibrahim.eldaghayes [at] vetmed.edu.ly Submitted: 02/05/2020 Accepted: 15/08/2020 Published: 02/09/2020 © 2020 Open Veterinary Journal
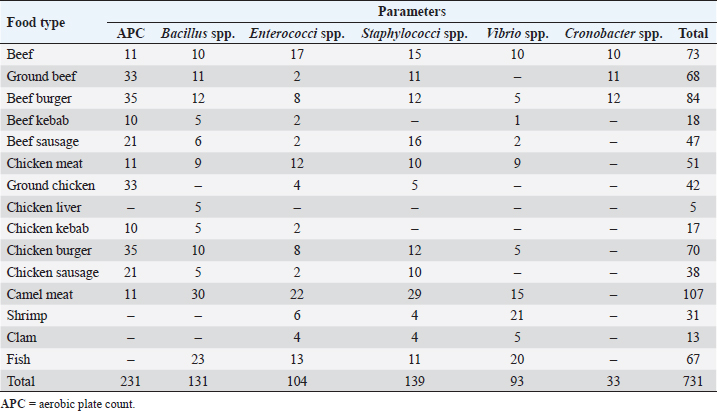
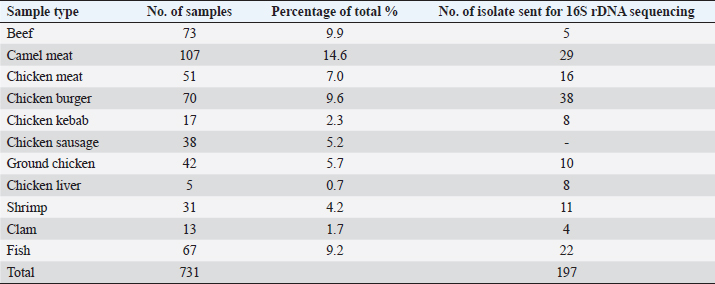
AbstractBackground: Whole muscle meat, meat products, and seafood contain different nutrients in adequate quantity providing a better environment for presence and replication of different microorganisms. There are underreported and inaccurate estimations of foodborne diseases due to the lack of effective surveillance systems in Libya. Aim: To determine the extent of microbiological contamination of whole muscle meat, meat products, and seafood. Methods: A total number of 731 samples of retail meat were collected from different stores in four cities in Libya. Samples were analyzed for aerobic plate count and subjected to microbiological enumeration and isolation techniques, followed by molecular identification by PCR and partial sequencing of 16S rDNA. Results: The results showed contamination of samples with enteric and spoilage bacteria. Fifteen genera of spoilage bacteria yielded 149 isolates which were detected and identified by PCR and partial sequencing of 16S rDNA as: Proteus spp., Provedencia spp., Raouttella ornithinolytical, Citrobacter spp., Enterobacter spp., Morganella morgi, Shewanella algea, Rhodobacter capsulatus, Listonella pelagia, Kluyvera spp., Pectobacterium spp., Brenneria spp., Klebsiella spp., Acintobacter radioresistens, and Pantoea spp. While for pathogenic bacteria, 143 isolates distributed among nine genera were identified by PCR and partial sequencing of 16S rDNA as: Bacillus spp., Escherichia spp., Shigella spp., Enterococci spp., Cronobacter spp., Staphylococci spp., Salmonella spp., Aeromonas spp., and Vibrio spp.. Many isolated bacteria are zoonotic bacteria with high importance for public health. Conclusion: Excessive handling and processing of meat and meat products seems to be one of the poorest microbiological qualities. These findings ought to be helpful in risk assessments and quality assurance of meat in order to improve food safety. Keywords: Libya, Meat products, Pathogenic bacteria, Spoilage bacteria, Whole muscle meat. IntroductionAny food, including meat and seafood, can be described as spoiled when it is considered unacceptable based on its sensory characteristics (Nychas et al., 2008). Meat is the most nutritive source of protein consumed by humans. The water content of meat, the main source of protein for humans, is high, corresponding to approximately 0.99 water activity, which is suitable media for microbial propagation (Rao et al., 2009). Factors that affect microbial growth in meat can be divided into intrinsic properties (physical and chemical) and extrinsic environmental factors (Johnson and McGowan, 1998). Microbial propagation in meat and meat products is influenced by many factors such as storage temperatures, oxygen availability, and moisture (Forest et al., 1985). Temperature is considered as the most important factor that can lead to spoilage of meat. Interruption in the meat cold chain may result in overgrowth of specific spoilage microorganisms and overproduction of biogenic amines (Nychas et al., 2008). Tissues from healthy animal are sterile; however, they can be contaminated by microorganisms during slaughter, dressing, and processing. These microorganisms mainly come from the animal skin and its intestinal tract; moreover, meat can get contaminated post-slaughtering by water, air, and soil, as well as from the workers and the equipment involved during processing (Lawrie, 1984; Ray and Bhunia, 2008). Recently, meat safety has become a significant concern, and inferences endure that challenges will proceed in the oncoming years. Meat-borne pathogens are subjected to heat (physical), organic acids (chemical), and nutritional stresses during meat processing (Yousef and Courtney, 2003). However, their elimination and/or inhibition remains a big hurdle to food processors. The most common bacteria isolated from fresh meat are bacteria of the genera Acinetobacter, Pseudomonas, Brochothrix, Flavobacterium, Psychrobacter, Moraxella, Staphylococci, Micrococci, lactic acid bacteria (LAB), and various genera of the Enterobacteriaceae. The survival and growth of these organisms are mainly influenced by the atmospheric composition of the meat. Furthermore, meat spoilage is most frequently caused by the following genera: Pseudomonas spp., Enterobacteriaceae, Brochothrix thermosphacta, and LAB (Pennacchia et al., 2011). Foodborne illnesses (with symptoms of diarrhea and vomiting) are not considered as serious health threats in Arab countries and typically go unreported. The lack of effective surveillance systems in Arabic countries, including Libya, is the main reason for underreporting and the consequent inaccurate estimation of the burden associated with foodborne diseases (FAO/WHO, 2005a, 2005b). No detailed studies have been carried to describe the development and final composition of the bacterial flora in fresh meat, meat products, and seafood in Libya. Nowadays, more advanced genotypic methods are accessible to investigate microbial taxonomy. Among those molecular techniques, the 16S rDNA analysis has been accepted as the most reliable method (Nour, 1998; Adams and Moss, 2002). This investigation, therefore, focused on detecting, isolating, and identifying the enteric and spoilage microorganisms by 16S rDNA sequencing in whole muscle food sold in retail markets in Libya, in order to emphasize the public health risk from the zoonotic bacteria and implications of consuming such contaminated meat or meat products and provides useful information where necessary to the general public and potential approaches for improving the quality assurance and creating awareness among the consumers. Table 1. Number and type of samples for each parameter.
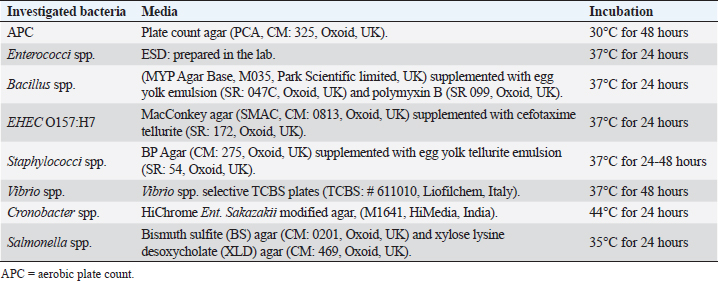
Materials and MethodsCollection and preparation of samplesA total of 731 individual retail samples, which included whole muscle meat, meat products, and seafood, were randomly collected from different cities in Libya (Janzour, Regdalin, Tripoli, and Tobruk); their numbers and types for each parameter are shown in Tables 1 and 2. The samples were collected in sterile polythene bags, packed in a carrier box containing ice packs and transported to the Food Hygiene and Control Laboratory, at the Faculty of Veterinary Medicine, University of Tripoli. Not all samples were tested for all parameters. The selection of parameters depended on the type of meat sample, the technology/processing received, and microbial ecology. Samples were subjected to the quantitative microbiological analyses, followed by molecular identification by PCR and partial sequencing of 16S rDNA. Decimal dilutions, culturing, and enumeration techniques of the samples were performed according to the methods described by the American Public Health Association (Downes et al., 2001). In brief, 25 g from each sample was aseptically transferred into a sterile stomacher bag (Seward Medicals, Rhymney, UK) and homogenized (Stomacher 400, Seaward Medicals, Rhymney, UK) with 225 ml of sterile peptone water 0.1% (w/v) (M: 0126, Park Scientific Limited, Northampton, UK) at 230 rpm for 2 minutes. Appropriate sample dilutions (100 µl) were spread onto plate count agar (PCA, CM: 325, Oxoid, Basingstoke, UK) plates for aerobic plate count (APC) and various selective agar plates for enumeration (quantitative) of different targeted microorganisms. The media used in the isolations of different microorganisms are listed in Table 3. Identification of isolated bacteria by PCR and partial sequencing of 16S rDNAGF-1 bacterial DNA extraction kit (Cat. # GF-BA-100, Vivantis, Malaysia) was used for all bacterial DNA extraction as described in a previous study (Naas et al., 2019). The 16S rDNA gene was amplified using the forward primer: S-D-Bact-0341-b-S-17 5′-CCTACGGGNGGCWGCAG-3′ and reverse primer: S-D-Bact-0785-a-A-21 5′-GACTACHVGGGTATCTAATCC-3′ (Herlemann et al., 2011). GF-1 Ambi Clean kit (Cat. # GF-GC-100, Vivantis, Malaysia) was used to get the amplified 16S rDNA PCR fragment (464 bp) from the gel (Azwai et al., 2016). The purified 16S rDNA amplicon was analyzed in cycle sequencing with Big Dye® Terminator v1.1 kit (AB Applied Bioscience, TECHNE, TC-512, Waltham, MA) and sequenced on a four-capillary ABI PRISM® 3130-Avant Genetic Analyzer at Istituto Zooprofilattico Sperimentale Della Lombardia e dell ‘Emilia Romagna (IZSLER) in Brescia, Italy. Sequences were assembled and edited using SeqMan module within the Laser gene package, (DNA Star Inc., Madison, WI). The obtained consensus sequences were subjected to BLAST search at both NCBI (http://www.ncbi.nlm.nih.gov/pubmed) and at 16S bacterial cultures Blast Server for the identification of prokaryotes (http://bioinfo.unice.fr/blast/). Table 2. Type and number of samples examined by conventional microbiological methods.
Table 3. Investigated bacteria and their isolation and identification procedures.
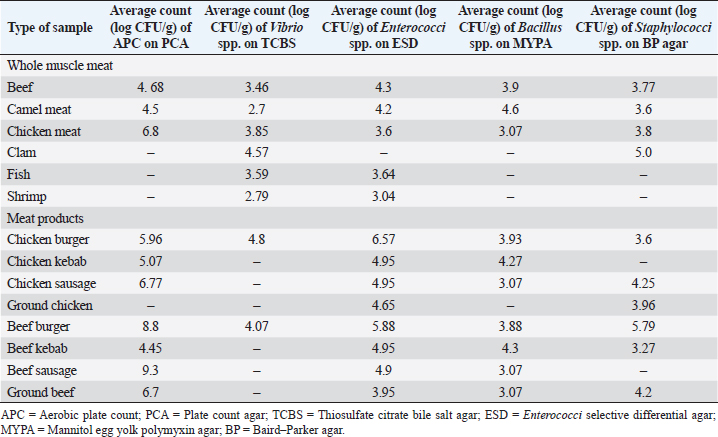
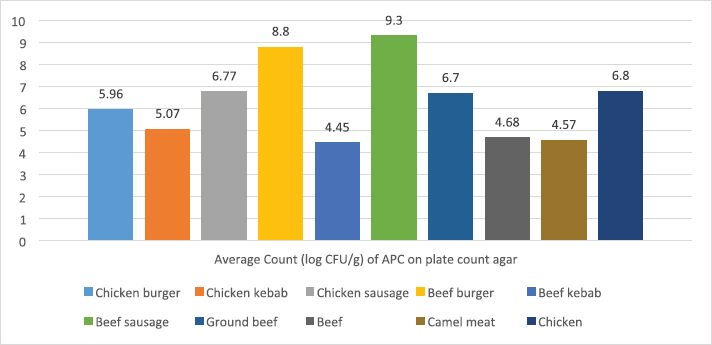
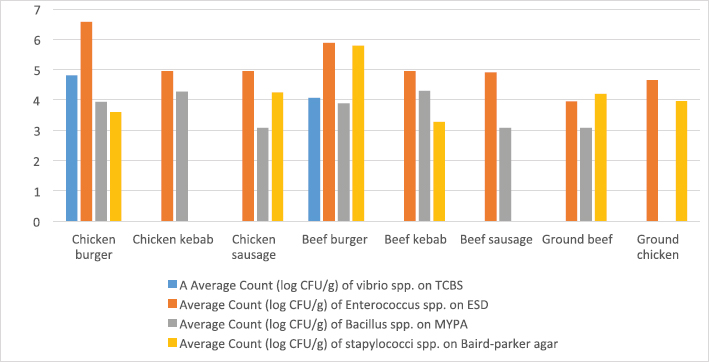
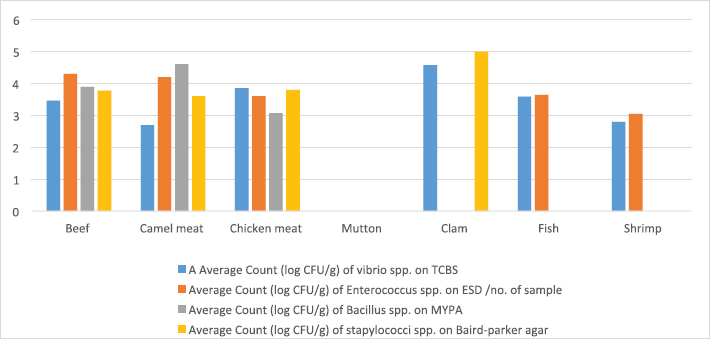
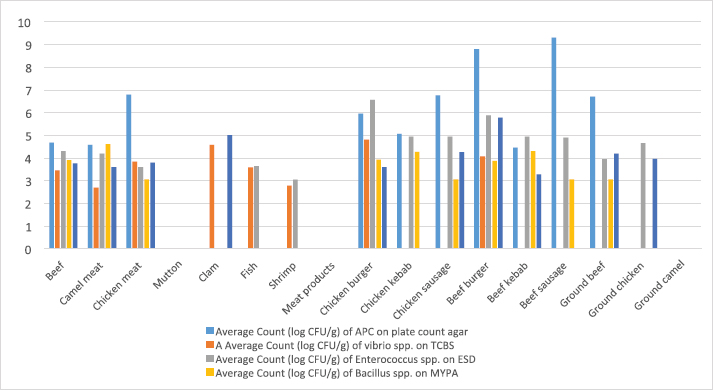
ResultsIsolation of spoilage and pathogenic microorganisms in whole muscle foodThe current investigation confirmed that the retail whole muscle meat, meat products, and seafood, obtained from different localities in Libya, were contaminated with enteric and spoilage bacteria. Table 4 shows the estimated average count of APC (log CFU/g) on PC agar and the average counts of Vibrio spp., Enterococci spp., Bacillus spp., and Staphylococci spp. in whole muscle meat and meat products on Thiosulfate citrate bile salt agar (TCBS), Enterococci selective differential agar (ESD), Mannitol egg yolk polymyxin agar (MYPA), and Baird–Parker agar (BP agar), respectively (Table 3). The results of the microbiological analysis of 231 whole muscle meat and meat products samples showed that the average APC counts ranged between 4.45 log CFU/g for beef kebab and 9.3 log CFU/g for beef sausage. While the results of the other parameters of 500 whole muscle meat and meat products samples showed that the average counts of Vibrio spp. were between 2.7 log CFU/g for camel meat and 4.8 log CFU/g for chicken burger, and the average counts of Enterococci spp. were between 3.04 log CFU/g for shrimp and 6.57 log CFU/g for chicken burger, the average counts of Bacillus spp. were between 3.07 log CFU/g for chicken, chicken sausage, beef sausage, and ground beef to 4.6 log CFU/g for camel meat, and the counts of Staphylococci spp. were between 3.27 log CFU/g for beef kebab and 5.79 log CFU/g for beef burger (Table 4) (Figs. 1–4). Table 4. Average count (log CFU/g) of APC and other tested bacteria in whole muscle food and meat products samples.
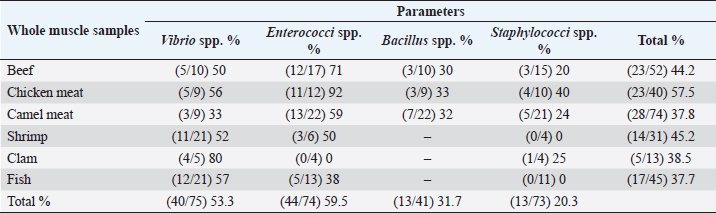
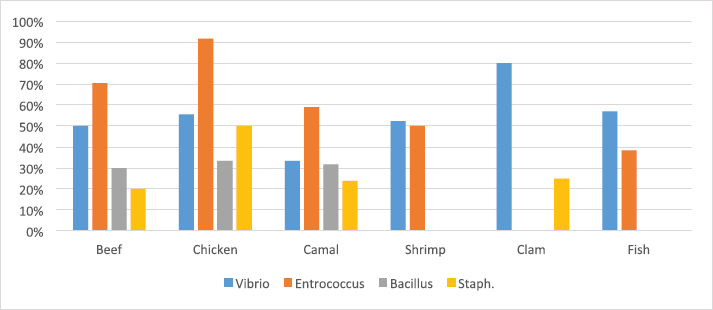
Table 5 and Figure 5 proved that the quality of whole muscle meat does not fulfill the set minimum limits of Libyan standards. The microbiological analysis of whole muscle meat for Vibrio spp., Enterococci spp., Bacillus spp., and Staphylococci spp. parameters showed that the incidence rates were the highest for chicken meat samples (56%, 92%, 33% and 50%, respectively), followed by beef samples (50%, 71%, 30% and 20%, respectively), while for camel meat samples the incidence rates were relatively low compared to chicken and beef samples. The incidence rate of Vibrio spp. in clam samples was the highest (80%), while in shrimp and fish samples it was 52% and 57%, respectively. Moreover, the incidence rate of Enterococci spp. in shrimp and fish was 50% and 38%, respectively. Staphylococci spp. frequency of 25% was only noted in clam samples.
Fig. 1. Average count (log CFU/g) of APC in whole muscle meat samples.
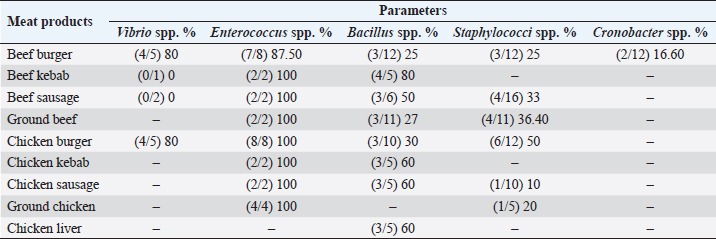
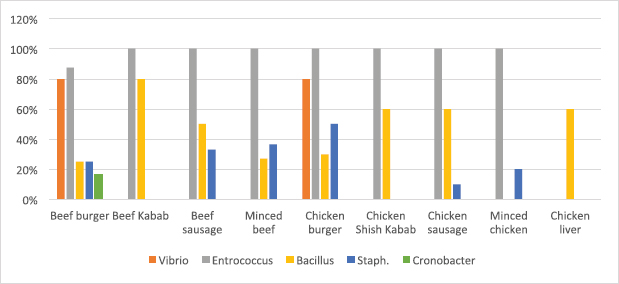
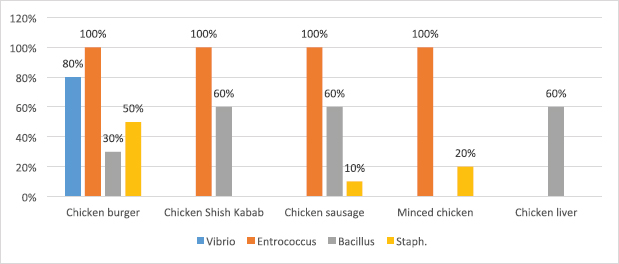
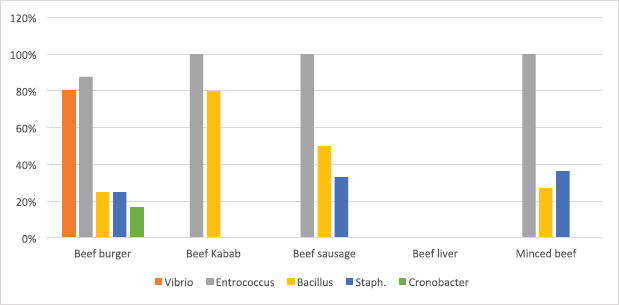
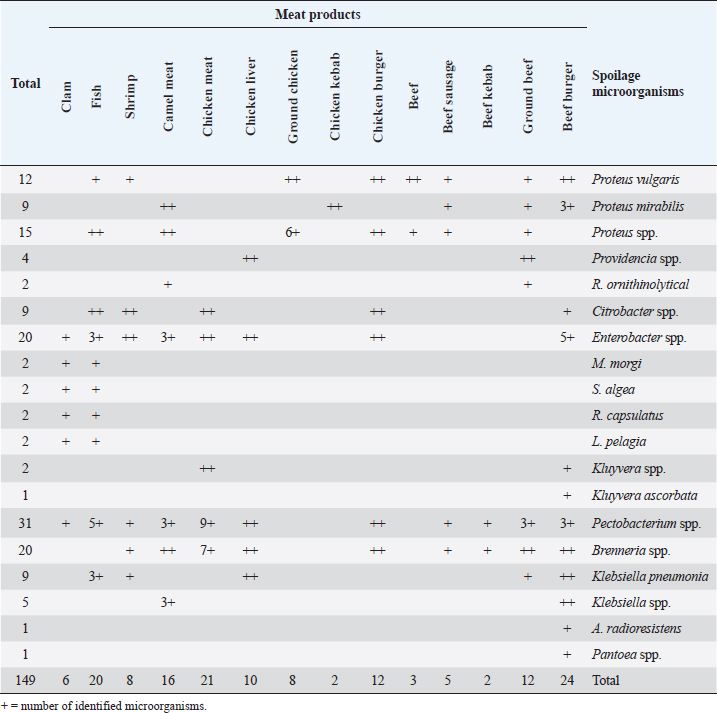
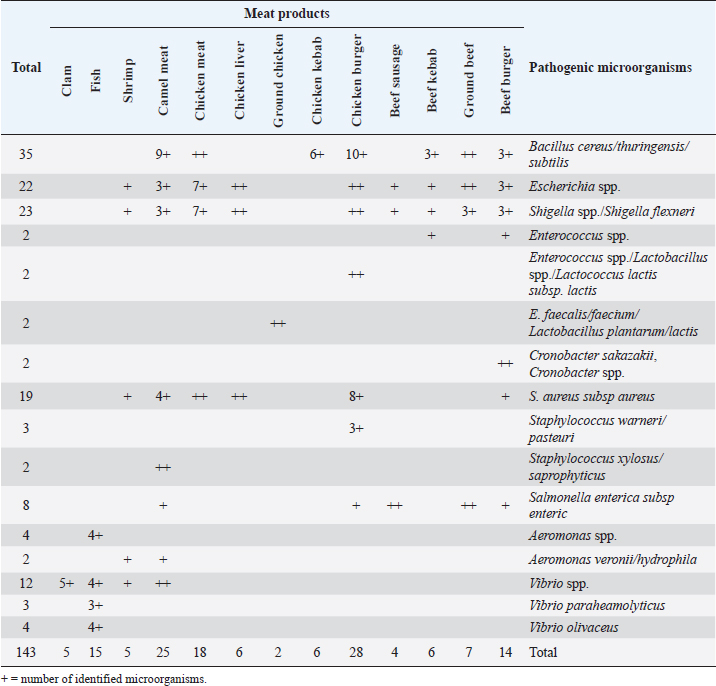
Fig. 2. Average count (log CFU/g) of tested bacteria in meat product samples. The findings of the microbiological analysis of meat product samples are shown in Table 6 and Figure 6. The incidence of Vibrio spp. in beef burger and chicken burger was 80%, while the incidence of Enterococci spp. was 100% in all tested meat products except in beef burger (87.5%). The detection of Bacillus spp. presence was high in beef kebab (80%) compared to beef burger (25%). In addition, the isolation rate of Staphylococci spp. in the chicken burger was 50% and was as low as 10% in chicken sausage, while the incidence of Cronobacter spp. in the beef burger was 16.6%. The findings of the microbiological analysis of chicken product samples and beef product samples are shown in Figures 7 and 8, respectively. Identification of spoilage and pathogenic microorganisms in whole muscle food samples by PCR and partial sequencing of 16S rDNAFifteen genera of spoilage bacteria yielded 149 isolates which were isolated from the whole muscle meat, meat products, and seafood samples and were identified by PCR and partial sequencing of 16S rDNA. These results are shown in Table 7 as Proteus spp., Provedencia spp., Raouttella ornithinolytical, Citrobacter spp., Enterobacter spp., Morganella morgi, Shewanella algea, Rhodobacter capsulatus, Listonella pelagia, Kluyvera spp., Pectobacterium spp., Brenneria spp., Klebsiella spp., Acintobacter radioresistens, and Pantoea spp. While for pathogenic bacteria, 143 isolates belonging to nine genera were identified as: Bacillus spp., Escherichia spp., Shigella spp., Enterococci spp., Cronobacter spp., Staphylococci spp., Salmonella spp., Aeromonas spp., and Vibrio spp. as shown in Table 8.
Fig. 3. Average count (log CFU/g) of tasted bacteria in muscle meat samples.
Fig. 4. Average count (log CFU/g) of APC and other tasted bacteria in whole muscle meat samples. The highest isolation rate of spoilage bacteria was 20.8% (31/149) isolates for Pectobacterium spp., while for enteric pathogens the highest isolation rate was 23.7% (34/143) isolates for Bacillus spp., followed by 16% (23/143) isolates for Shigella spp. DiscussionThe whole muscle meat samples from all locations yielded considerable bacterial loads. The presence of these organisms on whole muscle meat, meat products, and seafood has been explained as meat contains different nutrients in adequate quantity that can support the presence and multiplying of bacteria. Bacterial zoonotic diseases can be transferred from animals to humans in many ways and by different routes (Glaser et al., 1994). Zoonotic bacteria originating from food of animal origin can reach people through the direct fecal–oral route, contaminated animal food products, improper food handling, and inadequate cooking. Table 5. Percentage of suspected isolates yielded on different selective cultures in whole muscle meat samples.
Fig. 5. Percentage of suspected isolates yielded on different selective cultures in muscle meat samples. Table 6. Number of suspected isolates grown on different selective cultures in meat product samples.
Fig. 6. Percentage of suspected isolates yielded on different selective cultures in meat product samples.
Fig. 7. Percentage of suspected isolates yielded on different selective cultures in chicken product samples.
Fig. 8. Percentage of suspected isolates yielded on different selective cultures in beef product samples. Table 7. Number of spoilage microorganisms in whole muscle meat, meat products, and seafood identified by PCR and partial sequencing of 16S rDNA.
Microorganisms are the most important factor affecting the quality and safety of whole muscle meat and its products; therefore, it has been recommended that total viable counts (TVC) of meat spoilage should be less than 106 CFU/g (Panigrahi et al., 2006). Huge numbers of investigations have been conducted on the occurrence of spoilage and pathogenic bacteria in meat and its products worldwide (Holds et al., 2007; Kinsella et al., 2009). Moreover, the emergence of pathogens should not be a surprise as approximately 60%–70% of outbreaks and 40%–50% of reported cases of foodborne illness are caused by unknown etiologic agents (Sofos, 2008). The initial meat contamination flora is very heterogeneous with respect to microbial numbers and composition (Jay, 1996). This is in agreement with Ali et al. (2010), who reported that heavy bacteriological load was in meat carcasses with TVC ranging from 106 to 1010 CFU/g. The key objective of this investigation was to determine the microbiological quality (enteric and spoilage bacteria) of different whole muscle meat, meat products, and seafood sold in Libya. Table 8. Number of pathogenic microorganisms in whole muscle meat identified by PCR technique.
The results of the microbiological analysis of 231 whole muscle food samples showed that the average APC counts ranged between 4.45 log CFU/g for beef kebab and 9.3 log CFU/g for beef sausage. The high APC counts recorded in this study demonstrated the microbial diversity between the tested samples, condition of the markets, and the hygienic practice employed by meat producers, sellers, and butchers. This is an indication of recontamination during food handling and poor hygiene techniques used (Clarence et al., 2009). Similar results were reported by Yousuf et al. (2008), which pointed out that a bacterial count exceeding 5 log CFU/g in meat products is indicative of dangerous contamination. However, it has been proven that samples of ground meat contaminated with APC of 6 log CFU/g indicate a deterioration process with off-odor and reduction of shelf life and when contamination reaches 7 log CFU/g slime formation that has already happened. While the results of the other parameters of 500 whole muscle meat and meat products samples showed that the average counts of Vibrio spp. were between 2.7 log CFU/g for camel meat and 4.8 log CFU/g for chicken burger, the average counts of Enterococci spp. were between 3.04 log CFU/g for shrimp and 6.57 log CFU/g for chicken burger. These results were nearly similar to the incidence of Enterococci spp. in Japanese retail ready-to-eat raw fish 90 (45%) samples. Fifty-six strains were identified at the species level: Enterococcus faecalis, Enterococcus faecium, E. casseliflavus, E. gallinarum, E. phoeniculicola, E. raffinosus, E. saccharolyticus, and E. gilvus (Hammad et al., 2014). In addition, McGowan et al. (2006) found that among the meat items tested, enterococci were isolated from 95% of the chicken samples, 73% of the beef samples, 95% of the turkey samples, and 68% of the pork samples, and the predominant species was identified as E. faecalis from meat. Meanwhile, the average counts of Bacillus spp. were between 3.07 log CFU/g for beef kebab, beef sausage, and chicken sausage and 4.6 log CFU/g for camel meat, and the counts of Staphylococci spp. were between 3.27 log CFU/g for beef kebab and 5.79 log CFU/g for beef burger. According to Agriculture and Consumer Protection Department (Food and Agriculture Organization of the United Nations (FAO), 1991), the high log CFU/g of these viable bacteria is considered as a good indicator of the expected shelf life of meat that raises the chances of meat spoilage in a short time. Data in Table 5 and Figure 5 have demonstrated that the quality of whole muscle meat does not fulfill the set minimum limits of Libyan standards. The microbiological analysis of whole muscle meat for Vibrio spp., Enterococci spp., Bacillus spp., and Staphylococci spp. parameters showed that the incidence rates were the highest for chicken samples, followed by beef samples, while for camel samples the incidence rates were relatively low compared to chicken and beef samples. The incidence rate of Vibrio spp. in clam samples was the highest (80%), while in shrimp and fish samples it was 52% and 57%, respectively. Moreover, the incidence rate of Enterococci spp. in shrimp and fish was 50% and 38%, respectively. Staphylococci spp. Frequency of 25% was only noted in clam samples. The results of microbiological analysis of meat product samples are shown in Table 6 and Figure 6. The incidence of Vibrio spp. in beef burger and chicken burger was 80%, while the incidence of Enterococci spp. was 100% in all tested meat products, except in beef burger it was 87.5%. Detection of Bacillus spp. presence was high in beef kebab (80%) compared to beef burger (25%). In addition, the isolation rate of Staphylococci spp. in chicken burger was 50% and was as low as 10% in chicken sausage, while the incidence of Cronobacter spp. in beef burger was 16.6%. Another study in Benghazi city, Libya, reported microbial contamination of beef meat with bacterial strains which included Escherichia coli, Enterobacter spp., Citrobacter spp., Klebsiella spp., Proteus spp., Pseudomonas spp., and Salmonella spp. (Mansour et al., 2019). Yousuf et al. (2008) found some strains of Staphylococcus aureus, Salmonella spp., Shigella spp., Flavobacterium spp., and Vibrio spp. in local tiger shrimps and in giant water prawns from Bangladesh. The isolation of Enterobacter spp. could be due to poor environmental conditions such as dust and contaminated water used during slaughtering. Enterobacter spp. are also inhabitants of dairy products. Salmonella spp. (11.1%) is one of the most important pathogenic organisms of public health concerns in meat that may get contaminated through abusive handling (Okonko et al., 2009). A total of 292 isolates consisting of 15 genera of spoilage bacteria yielded 149 isolates which were isolated from whole muscle meat samples identified. The isolates were identified by 16S rDNA gene as Proteus spp., Provedencia spp., R. ornithinolytical, Citrobacter spp., Enterobacter spp., M. morgi, S. algea, R. capsulatus, L. pelagia, Kluyvera spp., Pectobacterium spp., Brenneria spp., Klebsiella spp., A. radioresistens, and Pantoea spp.. These findings are in agreement with an enormous number of researches in meat microbiology which have proved that spoilage is caused even by a small fraction of the so-called specific spoilage organisms that dominate (Nychas et al., 2008). The main sources of these microorganisms are found to be the animal’s intestinal tract and the environment in which the animal had a direct contact at some time before or during slaughter. Meat can be spoiled quickly under aerobic conditions (in the presence of an atmospheric air). Moreover, microbial spoilage occurs as a consequence of the growth and metabolic activities of spoiling bacteria. In most studies, the bacteria that dominate spoiled food have been considered as those responsible for spoilage and, in some studies, the criterion of microbiological acceptability (TVC reaching 7 log CFU/g) has been used to define spoilage (Zhang et al., 2012; Höll et al., 2016). The main defects associated with the spoilage of such products are off-odors and off-flavors, discoloration, and usually accompanied by gas production, slime formation, and a decrease in pH (Altajori and Elshrek, 2014). Cerveny et al. (2009) demonstrated that the types of microbes found in meat and meat products are greatly affected by storage conditions. They found that Pseudomonas spp., Moraxella spp., Psychrobacter spp., Acinetobacter spp., and Gram-negative psychrotropic members of the family Enterobacteriaceae are commonly present on chilled meat products; moreover, various species of Lactobacillus, Leuconostoc Corenobacterium, Weissella, and many other bacterial species. The most frequent bacteria to occur on fresh meat (fresh meat forms a significant proportion of meat marketing in Libya and other developing countries) are bacteria of the genera Acinetobacter, Pseudomonas, Brochothrix, Flavobacterium, Psychrobacter, Moraxella, Staphylococcus, and Micrococcus, LAB and various genera of the Enterobacteriaceae family. According to Pennacchia et al. (2011), meat spoilage is commonly caused by Pseudomonas spp., Enterobacteriaceae, B. thermosphacta, and LAB. Moreover, the genus Shewanella, which is also regularly found on meat, is a genus of microbes related to Pseudomonas spp.. Shewanella putrefaciens releases hydrogen sulfide causing meat spoilage. On the other hand, the pathogenic bacteria that have been identified by PCR technique in this study were 143 isolates distributed among nine genera which include Bacillus spp., Escherichia spp., Shigella spp., Enterococci spp., Cronobacter spp., Staphylococci spp., Salmonella spp., Aeromonas spp., and Vibrio spp. Tyson et al. (2017) evaluated several retail meat commodities for the presence of enterococci in USA from 2002 to 2014, and the contamination rate was 92%. ConclusionThe findings of our study indicate that the enteric pathogenic and spoilage bacteria were present predominantly. With regard to the microbial implication in handling, slaughtering, dressing, and processing, the distribution of meat and meat products may endanger human health. Escherichia coli and S. aureus are normal flora in humans and animals; so their presence in foods are indications of improper human handling. Hence, this investigation was taken up to determine the extent of microbial contamination of meat. Contamination prevention and/or reduction as in Hazards analysis critical control points system rather than the traditional way of inspection and testing of end-product to ensure meat safety is fundamentally required. As raw meats are heavily contaminated with microorganisms and are potential sources of foodborne infections, therefore raw meat handlers should receive education in food hygiene discipline. Meat and poultry processors and regulators should use process control techniques to ensure that the performance standards for meat and poultry are met. The presence of these organisms on whole muscle food could be attributed to the fact that meat contains an abundance of all nutrients required for the growth of bacteria in adequate quantity. Some groups recognized the presence of bacteria, especially Gram-negative organisms as an indicator of open-air meat spoilage, while others argued their assertion and considered the presence of a high number of background organisms as a pathogen-reduction strategy due to the organisms’ antagonistic effect against pathogenic bacteria and thus safe for meat quality. The high total plate counts recorded in this study showed the microbial diversity in these locations, condition of the market, and the hygienic practice employed by meat sellers and butchers determined the variation of bacterial contamination. AcknowledgmentsThe authors thank the Authority of Natural Science Research and Technology (ANSRT), as this research was part of a project titled “Genetic authentication of bacterial isolates from meat and milk products in Libya and establishing the FLBC” that was supported by a grant provided by ANSRT. The authors are grateful to Veronica Papini, a technician in IZSLER, Brescia, Italy, who performed the partial sequencing of 16S rDNA. Conflict of interestThe authors declare that there is no conflict of interest. ReferencesAdams, M.R. and Moss, M. 2002. Food microbiology, 3rd ed. London, UK: Royal Society of Chemistry Publication, pp: 27–30. Ali, N.H., Farooqui, A., Khan, A., Khan, A.Y. and Kazmi, S.U. 2010. Microbial contamination of raw meat and its environment in retail shops in Karachi, Pakistan. J. Infect. Dev. Ctries. 4, 382–388. Altajori, N.N. and Elshrek, Y.M. 2014. Microbiological quality of meat and meat products Marketed in Tripoli city, Libya: review. Int. Sci. Res. Rev. 1, 20–24. Azwai, S.M., Alfallani, E.A., Abolghait, S.K., Garbaj, A.M., Naas, H.T., Moawad, A.A., Gammoudi, F.T., Rayes, H.M., Barbieri, I. and Eldaghayes, I.M. 2016. Isolation and molecular identification of vibrio spp. by sequencing of 16S rDNA from seafood, meat and meat products in Libya. Open Vet. J. 6, 36–43. Cerveny, J., Meyer, J.D. and Hall, P.A. 2009. Microbiological spoilage of meat and poultry products. In Compendium of the microbiological spoilage, of foods and beverages. Food microbiology and food safety. Eds., Sperber, W.H. and Doyle, M.P. New York, NY: Springer Science and Business Media, pp: 69–868. Clarence, S.Y., Obinna, C.N. and Shalom, N.C. 2009. Assessment of bacteriological quality of ready to eat food (Meat pie) in Benin City Metropolis, Nigeria. African J. Microbiol. Res. 3, 390–395. Downes, F.P., Ito, K. and Association, A.P.H. 2001. Compendium of methods for the microbiological examination of foods. 4th Ed. Washington, DC, American Public Health Association. Food and Agriculture Organization of the United Nations (FAO). 1991. Guidelines for slaughtering, meat cutting and further processing. Available via: http://www.fao.org/DOCREP/004/T0279E/T0279E03.htm (Accessed 10 January 2020). Food and Agriculture Organization of the United Nations and World Health Organization (FAO/WHO). 2005a. FAO/WHO regional meeting on food safety for the Near East, Amman, Jordan. Practical actions to promote food safety. Final report. Available via: ftp://ftp.fao.org/es/esn/food/meetings/NE_report_en.pdf (Accessed 15 November 2019). Food and Agriculture Organization of the United Nations and World Health Organization (FAO/WHO). 2005b. FAO/WHO regional meeting on food safety for the Near East, Amman, Jordan. The impact of current food safety systems in the Near East/eastern Mediterranean region on human health. Available via: ftp://ftp.fao.org/es/esn/food/meetings/NE_wp2_en.pdf (Accessed 15 November 2019). Forest, D.C., Harold, D.A., Judge, B.A. and Robert, E.A. 1985. Different types of meat and meat product consumed by Nigerian. Principle of meat science. New York, NY: Freeman and Co. pop., pp: 4–178. Glaser, C.A., Angulo, F.J. and Rooney, J.A. 1994. Animal associated opportunistic infections among persons infected with the human immunodeficiency virus. Clin. Infect. Dis. 18, 14–24. Hammad, A.M., Shimamoto, T. and Shimamoto, T. 2014. Genetic characterization of antibiotic resistance and virulence factors in Enterococcus spp. from Japanese retail ready-to-eat raw fish. Food Microbiol. 38, 62–66. Herlemann, D.P., Labrenz, M., Jurgens, K., Bertilsson, S., Waniek, J.J. and Andersson, A.F. 2011. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. I. S. M. E. J. 5, 1571–1579. Holds, G., Pointon, A., Lorimer, M., Kiermeier, A., Raven, G. and Sumner, J. 2007. Microbial profiles of carcasses and minced meat from Kangaroos processed in South Australia. Int. J. Food Microbiol. 123, 88–92. Höll, L., Behr, J. and Vogel, R.F. 2016. Identification and growth dynamics of meat spoilage microorganisms in modified atmosphere packaged poultry meat by MALDI-TOF MS. Food Microbiol. 60, 84–91. Jay, J.M. 1996. Modern food microbiology, 5th ed. New York, NY: Chapman and Hall. Johnson, D.D. and McGowan, C.H. 1998. Diet/management effects on carcass attributes and meat quality of young goats. Small Rumin. Res. 28, 93–98. Kinsella, K.J., Prendergast, D.M., McCann, M.S., Blair, I.S., McDowell, D.A. and Sheridan, J.J. 2009. The survival of Salmonellae enteric serovar typhimurium DT 104 and total viable counts on beef surfaces at different relative humidities and temperatures. J. Appl. Microbiol. 106, 171–180. Lawrie, R.A. 1984. The preservation effect of smoke on meat. Elmford, NY: Meat Science Pergaman Press Inc. Maxwell House Fair view park, pp: 49–52. Mansour, A.M.A., Ishlak, A.M.M. and Haj-Saeed, B.A. 2019. Study of microbial contamination of beef meat in Benghazi markets. Libyan J. Basic Sci. 8, 38–48. McGowan, L.L., Jackson, C.R., Barrett, J.B., Hiott, L.M. and Fedorka-Cray, P.J. 2006. Prevalence and antimicrobial resistance of enterococci isolated from retail fruits, vegetables, and meats. J. Food Prot. 69, 2976–2982. Naas, H.T., Edarhoby, R.A., Garbaj, A.M., Azwai, S.M., Abolghait, S.K., Gammoudi, F.T., Moawad, A.A., Barbieri, I. and Eldaghayes, I.M. 2019. Occurrence, characterization, and antibiogram of Staphylococcus aureus in meat, meat products, and some seafood from Libyan retail markets. Vet. World, 12, 925–31. Nour, M. 1998. 16S±23S and 23S±5S intergenic spacer regions of lactobacilli: nucleotide sequence, secondary structure and comparative analysis. Res. Microbiol. 149, 443–448. Nychas, G.J.E., Skandamis, P.N., Tassou, C.C. and Koutsoumanis, K.P. 2008. Meat spoilage during distribution. Meat Sci. 78, 77–89. Okonko, I.O., Ogun, A.A., Adejoye, O.D., Ogunjobi, A.A., Nkang, A.O. and Adebayo-Tayo, B.C. 2009. Hazards analysis critical control points (HACCP) and Microbiology qualities of Sea-foods as affected by Handler’s Hygiene in Ibadan and Lagos, Nigeria. African J. Food Sci. 3, 35–50. Panigrahi, S., Balasubramanian, S., Gu, H., Logue, C.M. and Marchello, M. 2006. Design and development of a metal oxide based electronic nose for spoilage classification of beef. Sens. Actuators B Chem. 119, 2–14. Pennacchia, C., Ercolini, D. and Villani, F. 2011. Spoilage-related microbiota associated with chilled beef stored in air or vacuum pack. Food Microbiol. 28, 84–93. Rao, V.A., Thulasi, G. and Ruban, S.W. 2009. Meat quality characteristics of non-descript buffalos as affected by age and sex. World Appl. Sci. J. 6, 1058–1065. Ray, B. and Bhunia, A. 2008. Control by reduced water activity. In Fundamental food microbiology. Eds., Ray B. and Bhunia A., London, UK: CRC Press, pp: 385–390. Sofos, J.N. 2008. Challenges to meat safety in the 21st century. Meat Sci. 78, 3–13. Tyson, G.H., Nyirabahizi, E., Crarey, E., Kabera, C., Lam, C., Rice-Trujillo, C., McDermott, P.F. and Tate, H. 2017. Prevalence and antimicrobial resistance of enterococci isolated from retail meats in the United States, 2002 to 2014. Appl. Environ. Microbiol. 84, e01902–17; doi:10.1128/AEM.01902-17. Yousef, E. and Courtney, D. 2003. Basics of stress adaptation and implications in new generation foods. In Microbial stress adaptation and food safety. Eds., Yousef, A.E. and Juneja, V.K. Boca Raton, FL: CRC Press, pp: 1–30. Yousuf, A.H.M., Ahmed, M.K., Yeasmin, S., Ahsan, N., Rahman, M.M. and Islam, M.M. 2008. Prevalence of microbial load in shrimp, penaeus monodon and prawn, macrobrachium rosenbergii from Bangladesh. World J. Agric. Sci. 4, 852–855. Zhang, Q.Q., Han, Y.Q., Cao, J.X., Xu, X.L., Zhou, G.H. and Zhang, W.Y. 2012. The spoilage of air-packaged broiler meat during storage at normal and fluctuating storage temperatures. Poult. Sci. 91, 208–214. | ||
| How to Cite this Article |
| Pubmed Style Eshamah HL, Naas HT, Garbaj AM, Azwai SM, Gammoudi FT, Barbieri IL, Eldaghayes IM. Extent of Pathogenic and Spoilage Microorganisms in Whole Muscle Meat, Meat Products and Seafood sold in Libyan market. Open Vet. J.. 2020; 10(3): 276-288. doi:10.4314/ovj.v10i3.6 Web Style Eshamah HL, Naas HT, Garbaj AM, Azwai SM, Gammoudi FT, Barbieri IL, Eldaghayes IM. Extent of Pathogenic and Spoilage Microorganisms in Whole Muscle Meat, Meat Products and Seafood sold in Libyan market. https://www.openveterinaryjournal.com/?mno=124850 [Access: January 25, 2026]. doi:10.4314/ovj.v10i3.6 AMA (American Medical Association) Style Eshamah HL, Naas HT, Garbaj AM, Azwai SM, Gammoudi FT, Barbieri IL, Eldaghayes IM. Extent of Pathogenic and Spoilage Microorganisms in Whole Muscle Meat, Meat Products and Seafood sold in Libyan market. Open Vet. J.. 2020; 10(3): 276-288. doi:10.4314/ovj.v10i3.6 Vancouver/ICMJE Style Eshamah HL, Naas HT, Garbaj AM, Azwai SM, Gammoudi FT, Barbieri IL, Eldaghayes IM. Extent of Pathogenic and Spoilage Microorganisms in Whole Muscle Meat, Meat Products and Seafood sold in Libyan market. Open Vet. J.. (2020), [cited January 25, 2026]; 10(3): 276-288. doi:10.4314/ovj.v10i3.6 Harvard Style Eshamah, H. L., Naas, . H. T., Garbaj, . A. M., Azwai, . S. M., Gammoudi, . F. T., Barbieri, . I. L. & Eldaghayes, . I. M. (2020) Extent of Pathogenic and Spoilage Microorganisms in Whole Muscle Meat, Meat Products and Seafood sold in Libyan market. Open Vet. J., 10 (3), 276-288. doi:10.4314/ovj.v10i3.6 Turabian Style Eshamah, Hanan L., Hesham T. Naas, Aboubaker M. Garbaj, Salah M. Azwai, Fatim T. Gammoudi, Ilaria L. Barbieri, and Ibrahim M. Eldaghayes. 2020. Extent of Pathogenic and Spoilage Microorganisms in Whole Muscle Meat, Meat Products and Seafood sold in Libyan market. Open Veterinary Journal, 10 (3), 276-288. doi:10.4314/ovj.v10i3.6 Chicago Style Eshamah, Hanan L., Hesham T. Naas, Aboubaker M. Garbaj, Salah M. Azwai, Fatim T. Gammoudi, Ilaria L. Barbieri, and Ibrahim M. Eldaghayes. "Extent of Pathogenic and Spoilage Microorganisms in Whole Muscle Meat, Meat Products and Seafood sold in Libyan market." Open Veterinary Journal 10 (2020), 276-288. doi:10.4314/ovj.v10i3.6 MLA (The Modern Language Association) Style Eshamah, Hanan L., Hesham T. Naas, Aboubaker M. Garbaj, Salah M. Azwai, Fatim T. Gammoudi, Ilaria L. Barbieri, and Ibrahim M. Eldaghayes. "Extent of Pathogenic and Spoilage Microorganisms in Whole Muscle Meat, Meat Products and Seafood sold in Libyan market." Open Veterinary Journal 10.3 (2020), 276-288. Print. doi:10.4314/ovj.v10i3.6 APA (American Psychological Association) Style Eshamah, H. L., Naas, . H. T., Garbaj, . A. M., Azwai, . S. M., Gammoudi, . F. T., Barbieri, . I. L. & Eldaghayes, . I. M. (2020) Extent of Pathogenic and Spoilage Microorganisms in Whole Muscle Meat, Meat Products and Seafood sold in Libyan market. Open Veterinary Journal, 10 (3), 276-288. doi:10.4314/ovj.v10i3.6 |