| Research Article | ||
Open Vet. J.. 2023; 13(6): 690-696 Open Veterinary Journal, (2023), Vol. 13(6): 690–696 Original Research First detection and genetic characterization of chicken infectious anemia virus in the Mekong Delta, VietnamNguyen Duc Hien1,2, Dao Huyen Tran1, Tran Ngoc Bich1, Nguyen Phuc Khanh1 and Lam Thanh Nguyen1*1Faculty of Veterinary Medicine, College of Agriculture, Can Tho University, Can Tho, Vietnam 2Can Tho Sub-Department of Animal Health, Ministry of Agriculture and Rural Development, Can Tho, Vietnam *Corresponding Author: Lam Thanh Nguyen. Faculty of Veterinary Medicine, College of Agriculture, Can Tho University, Can Tho, Vietnam. Email: ntlam [at] ctu.edu.vn Submitted: 21/02/2023 Accepted: 07/05/2023 Published: 04/06/2023 © 2023 Open Veterinary Journal
AbstractBackground: Chicken infectious anemia (CIA) caused by the CIA virus (CIAV) is considered one of the most important immunosuppressive diseases affecting chickens and recently poses a great economic burden to the poultry industry worldwide. Aim: This study aims to identify the presence of CIAV in the Mekong Delta (MD), Vietnam, and to determine genotypes of CIAVs that are currently circulating in this area. Methods: Organ samples (spleen, liver, and thymus) of 144 chickens suspected with CIA from 47 poultry farms were collected. A total of 47 pooled samples, each containing 2–4 chickens from each farm, were tested for the presence of CIAV. Results: Twenty out of 47 pooled organ samples (pool of 2–4 chickens per farm) were positive for CIAV using polymerase chain reaction targeting the viral VP1 gene. The VP1 amplicons of eight representative CIAVs were subjected to sequencing and genetic characterization. Phylogenetic analysis based on partial VP1 gene sequence revealed that the CIAVs detected in the MD grouped into different genotypes of II, IIIa, and IIIc together with CIAVs previously detected in the northern Vietnam and other Asian countries. The phylogenetic analysis also confirmed that detected CIAVs genetically differed from vaccine strains. In addition, deduced amino acids of the VP1 identified several critical amino acid substitutions in the VP1 protein that are likely associated with the virulence of CIAV. Conclusion: This is the first report to detect and determine the genetic characterization of the circulating CIAVs in the MD. Therefore, this study provides an important understanding of the evolution of CIAVs and highlights the importance of implementing prompt control measures against CIAVs in the MD and Vietnam. Keywords: Chicken infectious anemia virus, Genetic characterization, Mekong delta, Vietnam. IntroductionChicken infectious anemia (CIA) is a highly infectious disease of young chickens. The disease is characterized by severe anemia, subcutaneous and muscular hemorrhages, thymic atrophy, and immunosuppression which increase the susceptibility, morbidity, and mortality of infected chickens up to 100% and 60%, respectively (Yuasa and Imai, 1986; Noteborn and Koch, 1995). CIA was first described in Japan in 1979 (Yuasa et al., 1979) and its etiological agent is the CIA virus (CIAV) which is the only member of the Gyrovirus genus from the Anelloviridae family (Todd et al., 1991; Rosario et al., 2017). The chicken is considered as the only recognized natural species for this virus and no other reservoir species have been defined (Pope, 1991). However, a recent study indicated that CIAV was also detected in canine serum (Liu et al., 2022). Therefore, CIA persists as an economically important disease-causing considerable health problems and economic losses to the poultry industry worldwide (Schat, 2009; Orakpoghenor, 2019) and a potential public health concern (Liu et al., 2022). The genome of CIAV is a circular single-stranded DNA molecule of approximately 2.3 kb which consists of three major partially overlapping open reading frames encoding for three viral proteins such as VP1 (51.6 kDa), VP2 (24 kDa), and VP3 (13.6 kDa) (Noteborn and Koch, 1995). Among the viral proteins, VP1 is the main target for most molecular studies since this is the major capsid protein and is associated with virulence and antigenicity of CIAV (Renshaw et al., 1996). In addition, the genetic classification of CIAV is mostly based on the VP1 gene. For example, CIAVs can be divided into five genetically different genotypes, namely I, II, III (with three sub-genotypes of IIIa, IIIb, and IIIc), IV, and V based on the nucleotide sequence of VP1 (Van Dong et al., 2019). Besides, CIAV can undergo genetic recombination to new genetic variants (Zhang et al., 2013). Several previous studies have demonstrated the presence of inter-genetic recombination of CIAV that leads to novel viral genotypes and/or sub-genotypes (Zhang et al., 2013; Li et al., 2017; Ou et al., 2018; Van Dong et al., 2019). In Vietnam, CIAV was first reported in northern provinces in 2015 via antibody detection in chicken serum (Trinh et al., 2015). Then, several other studies have been conducted to determine the epidemiological prevalence and molecular characterization of CIAV in northern Vietnam (Dao et al., 2018; Van Dong et al., 2019; Huynh et al., 2020); however, no study of CIAV in the Mekong delta (MD), Vietnam has been conducted. This study was the first to investigate the presence and genetic characterization of CIAV in the chicken population in the MD. Materials and MethodsStudy areas, period, and sample collectionFrom 2020 to 2021, 144 individual chickens were collected at 47 commercial chicken farms in the MD, Vietnam (Fig. 1). At each farm, two to four chickens displaying poor performance and weakness were selected for sample collection. Organ samples including spleen, liver, and thymus from the two to four chickens of each farm were pooled into one 1.5 ml tube to make individual pooled samples. Thus, a total of 47 pooled samples representing 47 chicken farms were tested for detection of CIAV in this study. None of the farms involved had a history of CIAV vaccine implementation. DNA extraction and polymerase chain reaction (PCR) assayThe total DNA was extracted from organ samples using a Tissue Viral Extraction Top PURE® kit (TBR, Vietnam) according to the manufacturer’s instructions. The extracted DNA was subsequently used as templates for PCR to detect viral DNA of CIAV's previous study (Yao et al., 2019). A pair of primers (forward: 5ʹ-AGCCGACCCCGAACCGCAAGAA-3ʹ; reverse: 5ʹ- TCAGGGCTGCGTCCCCCAGTACA-3ʹ) was used in PCR. The amplicon was expected at 1,390 base pair (bp) in the length of the partial VP1 gene. PCR was carried out using commercial Master Mix 2X BIO (Bioline, UK). Briefly, the reaction mixtures in a 24 µl volume containing 1 µl of forward primer (10 mM), 1 l of reverse primer (10 mM), 12 µl of master mix, 7 µl of nuclease-free water, and 3 µl template DNA. The amplification was conducted in a thermal cycler under the following condition initial denaturation of 94 µ for 4 minutes, followed by 40 cycles of denaturation at 94µ for 1 minute, annealing at 57µ for 1 minute, extension at 72µ for 2 minutes, and final extension at 72µ for 7 minutes. The resulting PCR products were electrophoresed using 1.5% agarose gel, strained by ethidium bromide (0.5 mg/ml), and then visualized under ultraviolet transillumination. The PCR products of interest were purified by using TopPURE® PCR/Gel ADN Purification kit (TBR, Vietnam).
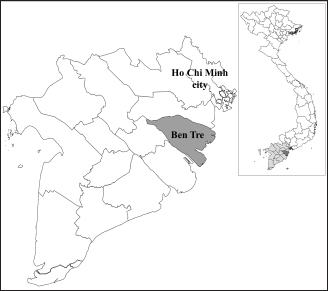
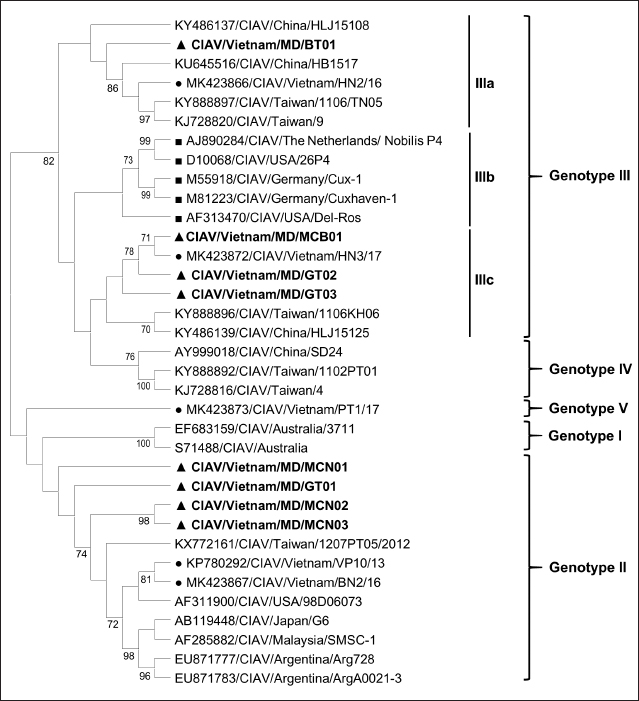
Fig. 1. Location of Ben Tre province where sampling was conducted. Ben Tre province (black) is situated in the MD (grey) on the Vietnam map. Sanger sequencing and phylogenetic analysisThe purified DNA products of positive pool samples were subjected to nucleotide sequencing in both directions by the Sanger method. The bidirectional electropherograms obtained from the Sanger method were assembled and manually inspected using the Bioedit program to ensure the quality of the sequencing. Then, sequence alignment was performed using Clustal W in MEGA X (Thompson et al., 1994; Hall, 2016). The phylogenetic relationship of the nucleotide sequences of 8 CIAV strains and other 24 CIAV reference strains obtained from Genbank were constructed using the maximum likelihood method with 1,000 bootstrap replication in MEGA X (Kumar et al., 2008). Recombination analysisPotential genetic recombination events in the VP1 gene of eight CIAV strains obtained in this study and reference CIAVs deposited in the Genbank were examined using nine algorisms supplemented in the Recombination Detection Program 5 (RDP 5), including RDP, GENECONV, MaxChi, Chimaera, BootScan, SiScan, Phylpro, LARD, and 3Seq with default settings (Martin et al., 2021). Only recombination events were defined if there is at least five of the nine methods with a p-value < 0.05 which is regarded as reliable. Ethical approvalAll experimental protocols were approved by the Institutional Animal Care and Use Committee of Can Tho University, Vietnam. Capture and dissection of animals were ethically performed following the guideline in accordance with the Regulation on Animal Experimentation of Can Tho University. ResultsDetection of CIAVsDetection of CIAV in collected samples was performed using PCR targeting the viral VP1 gene. Eight representative PCR amplicons at the expected size were shown in Figure 2 and defined as positive with CIAV (Table 1). A total of 20 pooled samples out of 47 were positive with CIAV, accounting for 42.55%. Genotyping of CIAVs using phylogenetic analysisRepresentative 8 out of 20 positive pool samples with CIAV were selected for sequencing and phylogenetic analysis. The result showed that these eight CIAVs were genetically grouped into two major genotypes II and III. Genotype II included four strains, namely CIAV/Vietnam/MD/MCN01, CIAV/Vietnam/MD/MCN02, CIAV/Vietnam/MD/MCN03, and CIAV/Vietnam/MD/GT01 (Fig. 3). genotype III also encompassed four strains of which three strains, CIAV/Vietnam/MD/MCB01, CIAV/Vietnam/MD/GT02, and CIAV/Vietnam/MD/GT03, belonged to sub-genotype IIIc and one strain, CIAV/Vietnam/MD/BT01, grouped into sub-genotype IIIa. In addition, the phylogenetic analysis also indicated that none of these CIAVs clustered into the sub-genotypes IIIb together with commercial vaccine strains such as Del-Ros, Nobilis P4, 26P4, Cux-1, and Cuxhaven-1. Amino acid substitutions in VP1Deduced amino acid sequences of the VP1 protein of eight current CIAVs were compared with previously published Vietnamese strains (HN2/16, HN3/17) and vaccine strains (Nobilis P4, 26P4). Five major variable amino acid substitutions were detected, namely V75I, I125L, K139Q, E144Q, and Q157E (Table 2). All of the CIAVs had threonine (T) and glutamine (G) at positions 89 and 141 of the VP1 sequence, respectively. Moreover, in the hypervariable region at positions 139 to 157 of the VP1 protein, five out of eight CIAVs in this study had glutamine (Q) at both positions 139 and 144 which are known to affect viral replication and infectivity. The remaining three CIAV strains possess lysine (K) at position 139 and glutamic acid (E) at position 144. In addition, CIAV/Vietnam/MD/BT01, CIAV/Vietnam/MD/MCN01, CIAV/Vietnam/MD/MCN02, CIAV/Vietnam/MD/MCN03, and CIAV/Vietnam/MD/GT01 have similar amino acid residues at the critical positions of 75, 89, 125, 139, 141, 144, and 157.
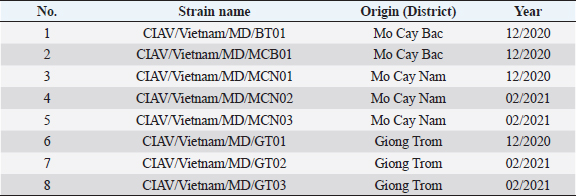
Fig. 2. PCR amplification of partial VP1 gene (1,390 bp) for detection of CIAV in the chicken organ samples collected in the MD, Vietnam. Lane M: 1 kbp DNA ladder, lane 1: negative control, lane 2: positive control, lane 3–10: eight representative positive samples of CIAVs. Information on these positive samples was indicated in Table 1. Table 1. Information of eight representative CIAV strains detected in this study.
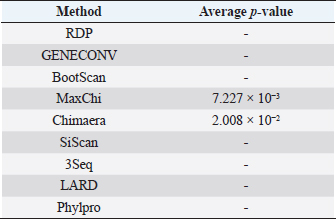
Fig. 3. Phylogenetic tree based on the partial sequence of VP1 gene of CIAV in the MD, Vietnam. ▲ CIAV strains detected in this study, ■ Vaccine strains, ● CIAV strains in northern Vietnam. The maximum likelihood method in MEGA X software was used to establish the phylogenetic tree (1,000 bootstrap replicates). Identification of genetic recombination in the VP1Recombination analysis using RDP5 software indicated that no inter-genetic recombination event was found among eight CIAV strains and other reference CIAVs. Only one out of eight CIAV strains, CIAV/Vietnam/MD/BT01, had significant recombination values (p-value < 0.05). However, recombination values were found only in two algorisms including Maxchi and Chimaera methods with a p-value of 7.227 × 10−3 and 2.008 × 10−2, respectively (Table 3). DiscussionCIA is considered one of the most economically important diseases for the poultry industry worldwide due to its highly infectious and immunosuppressive impacts. In Vietnam, CIA was first reported in 2015 in the northern area via antibody detection (Trinh et al., 2015), then several virological studies were subsequently conducted within the area. Most of these studies indicated that CIAVs circulate in chicken flocks at high prevalence, ranging from 47% to 62% (Dao et al., 2018; Van Dong et al., 2019; Huynh et al., 2020). Nevertheless, information about this virus in the MD remains unknown. Using PCR, 20 out of 47 (42.55%) pooled samples from 47 chicken farms in Ben Tre province, an administrative division in the MD (Fig. 1), were positive for CIAV. This result suggests that CIAVs currently persist and circulate in the chicken population across Vietnam with high prevalence. Prevalence of CIAVs in Vietnam was relatively equivalent to that of other Asian neighboring countries, namely China, Thailand, and Malaysia, which ranged from around 40% to 70% (Hailemariam et al., 2008; Chansiripornchai, 2016; Sun et al., 2022). It is noteworthy that we selected to collect samples from chickens with suspected CIAV infection signs such as anorexia, weakness, petechiation, and ecchymoses, lethargy. Therefore, the true prevalence in the herd might be lower than reported. On the other hand, real-time PCR or quantitative PCR is recommended for such epidemiological surveillance due to its higher sensitivity compared to conventional PCR, making it a more effective method for the detection of CIAV. However, it is important to highlight the spread and impacts of the disease on the poultry industry and the necessity to provide immediate control and prevention for CIAV in the MD and Vietnam where CIA vaccination is not currently available. Table 2. Comparison of deduced amino acids in the VP1 gene.
Table 3. Recombination statistics of CIAV/Vietnam/MD/BT01 in the VP1 resulted from RDP ver 5.0.
Genetic classification of CIAV has been divided into several different genotypes and sub-genotypes, mostly based on phylogenetic analysis of the VP1 nucleotide sequences (Van Dong et al., 2019). Our study indicated that the CIAV strains circulating in the MD belong to genotypes II, IIIa, and IIIc (Fig. 3) which are closely related to CIAVs previously reported in Vietnam (Dao et al., 2018; Van Dong et al., 2019; Huynh et al., 2020) and other Asian countries. It can be assumed that CIAVs in a country or geographical region tend to evolve locally within its genetic groups after being introduced from another country or region. Moreover, none of these strains clustered together with the vaccines’ genotype. Therefore, results from the phylogenetic analysis of the CIAV VP1 sequence suggested that the diversity of CIAV genotypes in the MD, and Vietnamese strains identified are closely related to the Chinese, Taiwanese, and Japanese CIAV strains. In this study, partial sequences of VP1 containing critical amino acid positions for biological properties of CIAVs were examined. Our results indicated a unique motif containing five amino acid substitutions in positions 75, 125, 139, 144, and 157 among the eight MD CIAV strains (Table 2). In particular, five out of eight CIAVs had amino acid motifs 75I and 125I which is similar to the motif pattern of the virulent strain HN2/16, suggesting that most of CIAVs circulating in the MD are pathogenic in chickens (Todd et al., 2002). The comparison between amino acid sequences showed that all of the CIAVs had amino acid motifs at the positions 75, 89, 125, 139, 141, 144, and 157 similar to either field strains of HN2/16 or HN3/17, and these motifs were also different from vaccine strains (Nobilis P4 and 26P4). This result also confirms that the MD CIAVs together with HN2/16 and HN3/17 could have a common ancestor and differ from vaccine strains. Recombination is one of the evolutionary processes that shape the architecture of viral genomes. Inter- or intra-genotypic recombination might generate new CIAV strains. In this study, none of the evidence of recombination events among the CIAV research sequences was detected (Table 3). As a DNA virus, gene recombination of CIAV is considered very low. In previous studies, recombination events in the CIAV genome have been reported in several countries including China (Tan et al., 2020; Zhang et al., 2022), Taiwan (Ou et al., 2018), and northern Vietnam (Van Dong et al., 2019; Huynh et al., 2020). In summary, the present study revealed the first detection of CIAV in the MD. The viral detection rate in chicken farms using conventional PCR was 42.55% which was relatively high and equivalent to that in previous studies in the North of Vietnam and other Asian countries. CIAVs detected in this study were determined to belong to genotypes II, IIIa, and IIIc. This study highlighted the circulation of CIAV and the necessity for further surveillance and strategies for the control of the CIA in the MD and Vietnam. AcknowledgmentsThe authors would like to thank the staffs of Ben Tre Sub-Department of Animal Health for joining field sampling. Conflict of interestThe authors declare that there is no conflict of interest regarding the publication of this article. Authors’ contributionsN.D.H. and L.T.N. designed the experimental procedures. D.H.T., N.P.K., and L.T.N. performed the experimental work. N.D.H., T.N.B., and L.T.N. conceptualized and designed the study. All authors read and approved the final manuscript. ReferencesChansiripornchai, N. 2016. Field study of seroconversion of three different commercial vaccines of chicken infectious anemia virus in Thailand. Thai J. Vet. Med. 46, 699–704. Dao, D., Cao, T., Vu, T., Nguyen, V. and Huynh, T. 2018. Prevalence of chicken infectious anemia virus (CIAV) circulating in Hanoi and surrounding provinces. Vietnam J. Agri. Sci. 16, 36–45. Hailemariam, Z., Omar, A.R., Hair-Bejo, M. and Giap, T.C. 2008. Detection and characterization of chicken anemia virus from commercial broiler breeder chickens. Virol. J. 5, 128. Hall, T. 2016. BioEdit v. 7.0. 5: Biological sequence alignment editor for Windows. Ibis Therapeutics a division of Isis pharmaceuticals, 2005. Huynh, L.T.M., Nguyen, G.V., Do, L.D., Dao, T.D., Le, T.V., Vu, N.T. and Cao, P.T.B. 2020. Chicken infectious anaemia virus infections in chickens in northern Vietnam: epidemiological features and genetic characterization of the causative agent. Avian Pathol. 49, 5–14. Kumar, S., Nei, M., Dudley, J. and Tamura, K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9(4), 299–306. Li, Y., Fang, L., Cui, S., Fu, J., Li, X., Zhang, H., Cui, Z., Chang, S., Shi, W. and Zhao, P. 2017. Genomic characterization of recent chicken anemia virus isolates in China. Front. Microbiol. 8, 401. Liu, Y., Lv, Q., Li, Y., Yu, Z., Huang, H., Lan, T., Wang, W., Cao, L., Shi, Y., Sun, W. and Zheng, M. 2022. Cross-species transmission potential of chicken anemia virus and avian gyrovirus 2. Infect. Genet. Evol. 99, 105249. Martin, D.P., Varsani, A., Roumagnac, P., Botha, G., Maslamoney, S., Schwab, T., Kelz, Z., Kumar, V. and Murrell, B. 2021. RDP5: a computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 7, veaa087. Noteborn, M.H. and Koch, G. 1995. Chicken anaemia virus infection: molecular basis of pathogenicity. Avian Pathol. 24, 11–31. Orakpoghenor, O. 2019. Chicken infectious anemia: emerging viral disease of poultry—an overview. Comp. Clin. Pathol. 28, 651–654. Ou, S.C., Lin, H.L., Liu, P.C., Huang, H.J., Lee, M.S., Lien, Y.Y. and Tsai, Y.L. 2018. Epidemiology and molecular characterization of chicken anaemia virus from commercial and native chickens in Taiwan. Transbound. Emerg. Dis. 65, 1493–1501. Pope, C.R. 1991. Chicken anemia agent. Vet. Immunol. Immunopathol. 30, 51–65. Renshaw, R.W., Soiné, C., Weinkle, T., O'Connell, P.H., Ohashi, K., Watson, S., Lucio, B., Harrington, S. and Schat, K.A. 1996. A hypervariable region in VP1 of chicken infectious anemia virus mediates rate of spread and cell tropism in tissue culture. J. Virol. 70, 8872–8878. Rosario, K., Breitbart, M., Harrach, B., Segalés, J., Delwart, E., Biagini, P. and Varsani, A. 2017. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch. Virol. 162, 1447–1463. Schat, K. 2009. Chicken anemia virus. Curr. Top. Microbiol. Immunol. 331, 151–183. Sun, H., Yu, S., Jiang, T., Yan, Z., Wang, D., Chen, L., Zhou, Q., Yin, L. and Chen, F. 2022. Molecular characterization of chicken infectious anaemia virus (CIAV) in China during 2020–2021. Avian Pathol. 52, 119–127. Tan, C., Wang, Z., Lei, X., Lu, J., Yan, Z., Qin, J., Chen, F., Xie, Q. and Lin, W. 2020. Epidemiology, molecular characterization, and recombination analysis of chicken anemia virus in Guangdong province, China. Arch. Virol. 165, 1409–1417. Thompson, J.D., Higgins, D.G. and Gibson, T.J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. Todd, D., Niagro, F., Ritchie, B., Curran, W., Allan, G., Lukert, P., Latimer, K., Steffens, W.D. and McNulty, M. 1991. Comparison of three animal viruses with circular single-stranded DNA genomes. Arch. Virol. 117, 129–135. Todd, D., Scott, A.N., Ball, N.W., Borghmans, B.J. and Adair, B.M. 2002. Molecular basis of the attenuation exhibited by molecularly cloned highly passaged chicken anemia virus isolates. J. Virol. 76, 8472–8474. Trinh, D.Q., Ogawa, H., Bui, V.N., Nguyen, T.T.H., Gronsang, D., Baatartsogt, T., Kizito, M.K., AboElkhair, M., Yamaguchi, S., Nguyen, V.K. and Imai, K. 2015. Development of a blocking latex agglutination test for the detection of antibodies to chicken anemia virus. J. Virol. Methods 221, 74–80. Van Dong, H., Tran, G.T.H., Van Nguyen, G., Dao, T.D., Bui, V.N., Huynh, L.T.M., Takeda, Y., Ogawa, H. and Imai, K. 2019. Chicken anemia virus in northern Vietnam: molecular characterization reveals multiple genotypes and evidence of recombination. Virus Genes 55, 643–653. Yao, S., Tuo, T., Gao, X., Han, C., Yan, N., Liu, A., Gao, H., Gao, Y., Cui, H. and Liu, C. 2019. Molecular epidemiology of chicken anaemia virus in sick chickens in China from 2014 to 2015. PLoS One 14, e0210696. Yuasa, N. and Imai, K. 1986. Pathogenicity and antigenicity of eleven isolates of chicken anaemia agent (CAA). Avian Pathol. 15, 639–645. Yuasa, N., Taniguchi, T. and Yoshida, I. 1979. Isolation and some characteristics of an agent inducing anemia in chicks. Avian Dis. 366–385. Zhang, M., Deng, X., Xie, Z., Zhang, Y., Xie, Z., Xie, L., Luo, S., Fan, Q., Zeng, T. and Huang, J. 2022. Molecular characterization of chicken anemia virus in Guangxi Province, southern China, from 2018 to 2020. J. Vet. Sci. 23, e63. Zhang, X., Liu, Y., Wu, B., Sun, B., Chen, F., Ji, J., Ma, J. and Xie, Q. 2013. Phylogenetic and molecular characterization of chicken anemia virus in southern China from 2011 to 2012. Sci. Rep. 3, 3519. | ||
| How to Cite this Article |
| Pubmed Style Hien ND, Tran DH, Bich TN, Khanh NP, Nguyen LT. First detection and genetic characterization of chicken infectious anemia virus in the Mekong Delta, Vietnam. Open Vet. J.. 2023; 13(6): 690-696. doi:10.5455/OVJ.2023.v13.i6.3 Web Style Hien ND, Tran DH, Bich TN, Khanh NP, Nguyen LT. First detection and genetic characterization of chicken infectious anemia virus in the Mekong Delta, Vietnam. https://www.openveterinaryjournal.com/?mno=144385 [Access: January 24, 2026]. doi:10.5455/OVJ.2023.v13.i6.3 AMA (American Medical Association) Style Hien ND, Tran DH, Bich TN, Khanh NP, Nguyen LT. First detection and genetic characterization of chicken infectious anemia virus in the Mekong Delta, Vietnam. Open Vet. J.. 2023; 13(6): 690-696. doi:10.5455/OVJ.2023.v13.i6.3 Vancouver/ICMJE Style Hien ND, Tran DH, Bich TN, Khanh NP, Nguyen LT. First detection and genetic characterization of chicken infectious anemia virus in the Mekong Delta, Vietnam. Open Vet. J.. (2023), [cited January 24, 2026]; 13(6): 690-696. doi:10.5455/OVJ.2023.v13.i6.3 Harvard Style Hien, N. D., Tran, . D. H., Bich, . T. N., Khanh, . N. P. & Nguyen, . L. T. (2023) First detection and genetic characterization of chicken infectious anemia virus in the Mekong Delta, Vietnam. Open Vet. J., 13 (6), 690-696. doi:10.5455/OVJ.2023.v13.i6.3 Turabian Style Hien, Nguyen Duc, Dao Huyen Tran, Tran Ngoc Bich, Nguyen Phuc Khanh, and Lam Thanh Nguyen. 2023. First detection and genetic characterization of chicken infectious anemia virus in the Mekong Delta, Vietnam. Open Veterinary Journal, 13 (6), 690-696. doi:10.5455/OVJ.2023.v13.i6.3 Chicago Style Hien, Nguyen Duc, Dao Huyen Tran, Tran Ngoc Bich, Nguyen Phuc Khanh, and Lam Thanh Nguyen. "First detection and genetic characterization of chicken infectious anemia virus in the Mekong Delta, Vietnam." Open Veterinary Journal 13 (2023), 690-696. doi:10.5455/OVJ.2023.v13.i6.3 MLA (The Modern Language Association) Style Hien, Nguyen Duc, Dao Huyen Tran, Tran Ngoc Bich, Nguyen Phuc Khanh, and Lam Thanh Nguyen. "First detection and genetic characterization of chicken infectious anemia virus in the Mekong Delta, Vietnam." Open Veterinary Journal 13.6 (2023), 690-696. Print. doi:10.5455/OVJ.2023.v13.i6.3 APA (American Psychological Association) Style Hien, N. D., Tran, . D. H., Bich, . T. N., Khanh, . N. P. & Nguyen, . L. T. (2023) First detection and genetic characterization of chicken infectious anemia virus in the Mekong Delta, Vietnam. Open Veterinary Journal, 13 (6), 690-696. doi:10.5455/OVJ.2023.v13.i6.3 |