| Research Article | ||
Open Veterinary Journal, (2024), Vol. 14(1): 53-69 Original Research Summer mortality syndrome bacterial pathogens in farmed Nile tilapia (Oreochromis niloticus)Fawzy I. Magouz1, Eman Moustafa Moustafa2*, Etab M. Abo-Remela3,4, Marwa R. Halawa5, Passant M. Barakaat1 and Amira A. Omar21Fish Nutrition, Faculty of Agriculture, Kafrelsheikh University, Kafr El-Sheikh, Egypt 2Fish Diseases and Management Department, Faculty of Veterinary Medicine, Kafrelsheikh University, Kafr El-Sheikh, Egypt 3Bacteriology, Mycology and Immunology Department, Faculty of Veterinary Medicine, Kafrelsheikh University, Kafr El-Sheikh, Egypt 4Biology Department, Faculty of Science, Taibah University, Medina, Saudi Arabia 5Central Diagnostic and Research Laboratory, Faculty of Veterinary Medicine, Kafrelsheikh University, Kafr el-Sheikh, Egypt *Corresponding Author: Mousa E. Ahmed. Zagazig Provincial Laboratory, Animal Health Research Institute, Zagazig, Egypt. Email: emantarek2002 [at] yahoo.com Submitted: 01/10/2023 Accepted: 15/12/2023 Published: 31/01/2024 © 2024 Open Veterinary Journal
ABSTRACTBackground: The high summer mortality in many fish farms, which had detrimental economic and social implications, was a serious challenge that the fish industry had to deal with. Aim: With an examination of the most effective antibiotic, the ongoing research was intended to shed light on the identification of the main bacterial pathogens associated with the summer mortality syndrome in the diseased farmed Nile tilapia. Methods: Six hundred dead Nile tilapia samples that had suffered from summer mortality were collected from several fish farms between May and October of 2022. The gathered fish displayed hemorrhagic areas on the skin, scale detachment, fin degeneration, erosions, skin ulcers, and corneal opacity with unilateral and/or bilateral exophthalmia. The most prominent internal appearance was swelling of the internal organs with sanguineous ascetic fluid. Results: There were 225 bacterial isolates found. Six species were identified through phenotypic and biochemical analysis; they were Aeromonas, Vibrio, Streptococcus, Pseudomonas, Enterococcus, and Edwardsiella spp., in descending percentage, respectively. Aeromonas spp., Vibrio spp., and Streptococcus spp. were the three most frequent isolated bacterial pathogens. The identification of Aeromonas hydrophila, Vibrio spp., and Streptococcus iniae, the three most common bacterial isolates, was confirmed by molecular analysis by polymerase chain reaction. Most of the tested strains were found to be responsive to Ciprofloxacin (CIP), Gentamicin (CN), and Chloramphenicol (C) but resistant to Amoxicillin (AMX), according to an antibiotic sensitivity test. Conclusion: The three most dangerous common bacterial infections discovered during mass-farmed tilapia summer mortality are A. hydrophil a, Vibrio sp., and S. iniae. This makes it clear that high water temperatures may raise the possibility of bacterial infections, which could cause widespread tilapia mortality and substantial financial losses. Therefore, it is crucial to maintain a beneficial fish culture, environment, and husbandry practices to enhance the tilapia-rearing environment and lessen the virulence of the disease. Isolated bacterial strains showed low levels of resistance to AMX but were vulnerable to CIP, CN, and C. Keywords: Aeromonas hydrophila, Nile tilapia, Streptococcus iniae, Summer mortality, Vibrio sp. IntroductionAquaculture is now one of the most crucial methods for supplying the market because natural fisheries have been badly depleted and are no longer a reliable source of protein for humans (Mohamed et al., 2017; Adelekea et al., 2021). According to Reda et al. (2021), Egypt is the world’s third-biggest tilapia manufacturer and the aquaculture leading supplier in Africa. In addition to serving as an excellent provider of protein, tilapia is regarded as one of the essentials to ensuring the security of food, especially in densely inhabited nations like Egypt (Hussain et al., 2011; Abbas et al., 2017; El-Sayed, 2019). Moreover, many young individuals find employment in this sector as a means of support (Dickson et al., 2016). Extensive fish farming is unfortunately plagued by plenty of issues, such as diseases, that discourage farmers because of the financial penalties (Eltholth et al., 2015). Summer high mortality in several aquaculture facilities and the Nile Delta, which has had detrimental economic and social impacts, has been a key concern for this industry since 2013 (Fathi et al., 2017; Ali et al., 2020). The agents responsible for these outbreaks have been traced in several trials. Studies have revealed a number of variables, such as bacterial, fungal, viral, and parasitic causes, that may contribute to tilapia mortality (Yambot, 1998; Ye et al., 2011; Eissa et al., 2015; Fathi et al., 2017; Nicholson et al., 2020; Youssuf et al., 2020). Under stressed conditions, farmed Nile tilapia is vulnerable to several pathogenic microorganisms (Shaheen et al., 2013; Eissa et al., 2015). The most common cause of the large annual losses in the fish farming industry, which are projected to be worth billions of dollars (Elsheshtawy et al., 2019; Algammal et al., 2020), has been identified as bacterial infections. When water temperatures rise, opportunistic microorganisms in particular represent a serious threat to aquaculture (Pridgeon and Klesius, 2012). The potential for a bacterial infection to result in extensive fish mortality is a complex situation, and many aspects linked to the environment, host, and microbes may collaborate to show the progression of diseases (Harvell et al., 1999). Moreover, disease outbreaks may be sparked by environmental factors including temperature, salinity, oxygen level, and pH, especially throughout intensive aquaculture systems (Eissa et al., 2013). Due to the serious practices involved in fish farming, infectious illnesses constitute the main issue in aquaculture facilities and have a significant financial impact (Bulfon et al., 2015). Aeromonas veronii biovar sobria infection was linked to widespread deaths of cultured tilapia in the El-Sharkia governorate (Eissa et al., 2015; Elsheshtawy et al., 2019; Reda et al., 2021). However, Fathi et al. (2017) linked these fatalities to tilapia lake virus infection. According to Fang et al. (2004), Aeromonas is a dangerous disease that has the potential to spread internationally and produce epidemics in fish farming with extraordinarily high fatality rates. Aeromonas spp. are facultatively anaerobic, Gram-negative, oxidase-positive bacteria, according to Stratev and Odeyemi (2016). Aeromonads can be found in several kinds of foods, including fish, prawns, mussels, milk, meat, and vegetables, according to Neyts et al. (2000). They are frequently found in the aquatic environment. In tilapia farming, Aeromonas hydrophila has been found to be the most abundant species Hassan et al. (2017). Several Vibrio species are also widely recognized for their capability to lead to serious fish diseases. The demise of the population occurs when mortality in raised fish happens frequently during the earliest larval stages with rapid incidence (Thompson et al., 2004). The majority of the nonspore-producing, facultative, Gram-negative members of this family, Vibrionaceae, are typically distinguished by comma-shaped rods. According to Gobarah et al. (2023), the most typical heterotrophic microorganisms in the seawater ecosystem are Vibrio species. They are found in large numbers in brackish and/or coastal seawaters and have been isolated from fish, shellfish, and saltwater. One of the bacterial infections in aquaculture that is most frequently reported, streptococci are a nonsporing, facultatively anaerobic, Gram-positive bacterial pathogen that is regarded as a difficult problem in Egypt’s tilapia and catfish production (Liamnimitr et al., 2017). Many Streptococcus species can cause fish mortality; Streptococcus iniae, Streptococcus agalactiae, and Streptococcus dysgalactiae (Chen et al., 2007; Abdelsalam et al., 2010 and 2013). The most prevalent fish pathogen with a significant detrimental effect on aquaculture is S. iniae (Acar et al., 2015). It has a lot of virulence genes, which allow it to get inside fish and other aquatic creatures, reproduce, and evade their immune systems (Vazirzadeh et al., 2019). The goal of the ongoing research was to spotlight light on the identification of main bacterial pathogens involved in summer mortality syndrome isolated from diseased farmed Oreochromis niloticus throughout the summer season in the province of Kafr El-Sheikh, Egypt, and to investigate the antibiotic sensitivity test for the most predominant ones. Materials and MethodsThis investigation was executed during the summer months of 2022 at the Fish Diseases Laboratory, Veterinary Medicine Faculty at Kafrelsheikh University, Egypt. Samples collection and gross examinationFrom May to October 2022, 600 (at a rate of 100 fish/ month) clinically decrepit Nile tilapia, O. niloticus fish, with varying sizes (30–300 g), were randomly sampled alive or recently dead from multiple tilapia farms in Kafr El-Sheikh governorate. Fish farms, where samples of fish were gathered, suffered from severe summer mortalities with gross lesions of anorexia, surface swimming, hemorrhagic patches over the skin, scale detachment, fin degeneration and erosions, skin ulcers, corneal opacity with unilateral and/or bilateral exophthalmia. Samples of fish were collected, placed in a sterilized, sturdy polyethylene bag with pumped water and compressed oxygen in half of the bag’s volume, and immediately transported, while surviving, to the bacteriological section at the Central Diagnostic Laboratory, Fac. Vet. Med. There, the fish samples were kept alive with proper preparation and indefinite aeration using an electronic aerator until the examination (Hamouda et al., 2019). According to Eissa (2016), O. niloticus underwent a clinical examination to document any postmortem lesions and evident clinical anomalies. Isolation of the bacteria responsible for tilapia farms’ summer mortalitySkin, liver, kidney, spleen, and intestine tissue specimens from the captured tilapia were aseptically collected for bacteriological examination. Tissue specimens were first pre-enriched over Tryptic Soy Broth (Difco, Detroit, MI) and the incubation process was for 18–24 hours at 28°C using a sterilized loop. Next, they were dispersed out of tryptic soy agar (TSA) and kept for 48 hours at 28°C following preliminary enrichment on tryptic soy broth (TSB) (Austin and Austin, 2012). Pure isolated colonies over TSA have been then striped on selective diagnostic agar medium, following Markey et al. (2013), where Rimler Shotts (RS) agar for Aeromonas sp., Pseudomonas sp., and Edwardsiella sp.; Thiosulphate Citrate Bile salts Sucrose agar (TCBS) (Oxoid, UK) for Vibrio spp., Pseudomonas-F-agar base media for Pseudomonas fluorescens, blood agar, Edwards Medium (modified) (Oxoid, UK) with the addition of 5%–7% bovine blood and colistin sulfate 5 ml/l per manufacturer’s instructions and Streptococcus selective agar was used to test for Streptococuss sp. for 24 hours at 28°C–37°C. In addition, to isolate Enterococci bacteria, specimens of tissues were pre-enriched on Enterococcus M broth medium and selectively distributed on BBLTM Enterococcosel Agar (EA) before being incubated for a night at 37°C. The classification of colonies of bacteria was done by Reynolds (2011). On the same media, individual colonies of well-differentiated single bacterial colonies were produced, and these were subsequently identified. Gram’s stain was applied to the smears from the presumed pure colonies, and they were then examined using an oil immersion lens. A single representative colony was taken from each type of agar media, purified onto it, and then inoculated into brain heart infusion broth (Sigma-Aldrich). This broth included 15% glycerol and was aerobically incubated at 28°C for 24 hours before being conserved at −80°C. Identification of the bacteria responsible for tilapia farms’ summer mortalityThe bacterial isolates underwent phenotypic identification as explained previously (Schaperclaus et al., 1992; Bergey’s, 1994). It was tested for motility and Gram staining (Cruickshank et al., 1975). Furthermore, every isolate was biochemically verified using the standards outlined by Kreig and Holt (1984) and MacFaddin (2000). Biochemical examination tests such as catalase, oxidase, voges proskauer, methyl red, indole, triple sugar iron, citrate utilization, gelatin liquefaction, urease, starch hydrolysis, arginine, and lysine decarboxylase as well as sugar utilization and oxidation-fermentation tests were carried out for the recognition as well as bacterial strain distinguishing characters and the outcomes were contrasted with the reference strains for the identified bacteria (obtained from the Fish diseases department, Fac. Vet. Med., Kafrelsheikh University, Egypt) (Quinn et al., 2002; Nicky, 2004 and Markey et al., 2013) (Table 1). Molecular identification by polymerase chain reaction (PCR)DNA extraction for A. hydrophila and Vibrio sp. using QIA amp kitThe QIA amp kit (QIAamp: Qiagen Inc., USA) was used for the DNA extraction process. Correctly, 1 ml of the suspension of the young cell was centrifuged at 10,000 g for ten minutes at 4°C. The pellet of cells was then combined with 600 μl of guanidine hydrochloride buffer (pH 8.0), kept at ambient temperature for 30 minutes, and then centrifuged again at 10,000 g for 10 minutes at 4°C. 500 l of the supernatant was relocated to a different tube, blended with 100% ice-cold ethanol, and centrifuged at 13,000 × g for 20 minutes at 4°C. After washing the pellet with 95% and 90% ethanol, respectively, and discarding the supernatant, it was centrifuged at 10,000 g for 1 10 minutes at 4°C. Before being utilized as a PCR template, the pellet was reconstituted in 50 l of molecular-grade water and kept at −20°C (Haldar et al., 2005). DNA extraction of Streptococcus speciesBoiling techniques were used to separate the isolates’ genomic DNA from a pure culture. From the slants of the previously retrieved bacteria, three to five characteristic colonies of the same morphological type were extracted and they were enriched for 18 hours at 37°C in a tube containing 2 ml of TSB. A microcentrifuge tube containing 1 ml of the enhanced bacterial culture was centrifugated at 8,000 × g for 2 minutes, after which the sediment was homogenized with nuclease-free water and heated at 95°C for 15 minutes. After centrifuging the boiled lysates, the supernatant was employed as a DNA template. To identify Streptococcus, all DNA samples were brought to the Central Diagnostic and Research Laboratory. Table 1. The primers were used for the amplification of different associated genes for isolated summer mortality bacteria.
PCR identification system by using primer sequencesTable 1 introduces the primers that were used to identify the 16srRNA gene in A. hydrophila, the Vibrio sp., and the Sin gene in several Streptococcus species. PCR protocol for the isolatesAccording to the instructions on the Emerald Amp GT PCR master mix (Takara, Cat. PR310A) kit, the conditions for cycling for each gene were set up. The process of amplification was carried out on an Eppendorf MasterCyclerR (Eppendorf AG, Hamburg, Germany) in a total reaction volume of 25 μl, which contained 12.5 μl of the EmeraldAmp GT PCR Master Mix, 1.5 μl of each forward and reverse primer, 4.5 μl of molecular biology grade water, and 5 μl of the test DNA. The amplification conditions for A. hydrophila (using primers targeting 625 bp) and Vibrio sp. (using primers targeting 663 bp) were; 5 minutes of primary denaturation at 94°C, 35 cycles of secondary denaturation for 30 seconds at 94°C, annealing (16S rRNA PCR 40 seconds at 56°C for Vibrio while 40 seconds at 50°C for16S rRNA for A. hydrophila and extension at 72°C for 45 seconds (16S rRNA and 1 minutes). A final extension was adjusted for 10 minutes. The amplification conditions for S. iniae (using primers targeting 300 bp) were performed on a Thermal Cycler (Master cycler, Eppendorf, Hamburg, Germany). PCR assays were adopted in a 25 μl reaction mixture consisting of 12.5 μl of 2× MasterMix (intron), 1.5 μl of 20 pmol of each oligonucleotide primer, 5 μl of DNA template, and 4.5 μl Milli-Q water. The amplification profile consisted of an initial denaturation at 95°C for 15 minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 50°C for 30 seconds, and extension at 72°C for 30 seconds, with a final extension of 72°C for 10 minutes. Following amplification, the amplified DNA fragments were analyzed by 1.5% agarose gel electrophoresis (Applichem, Germany, GmbH). Lastly, the gel was photographed and seen under a UV transilluminator after being stained with ethidium bromide. To calculate the fragment sizes, a 100 bp plus DNA ladder (Qiagen, Germany, GmbH) was utilized (Moustafa et al., 2021). Antibiotic susceptibility testing (Antibiogramme)The retrieved Aeromonas, Vibrio, and Streptococcus strains were subjected to an in-vitro antibiotic sensitivity test to determine their susceptibility to various antibiotics utilizing the disc diffusion method (Koneman et al., 1997). The following antimicrobial agents were used: amoxicillin (AMX10 Mg), ciprofloxacin (CIP5 Mg), gentamicin (CN) (GEN10 Mg), and streptomycin (S). At least 4–5 colonies of each isolated strain were seeded into Mueller Hinton broth tubes, which were then cultured for 24 hours at 37°C. One ml of the infected broth was then applied to the Mueller Hinton agar plates, and they were incubated at 37°C for 24 hours. The data were interpreted by CLSI (2016). Ethical approvalAll procedures, handling of animals, and testing procedures employed in the current research were completed in compliance with the applicable standards and rules issued by Kafrelsheikh University’s animal ethics committee. ResultsClinical and postmortem examinationAnorexia, lethargy, surface swimming, body hemorrhages, scale detachment, fin degeneration and erosions, and corneal opacity with unilateral and bilateral exophthalmia (popeyes) were all symptoms of naturally infected O. niloticus (Fig. 1A–E). While this was going on, a postmortem evaluation of the animal revealed an accumulation of the abdominal cavity’s sanguineous ascitic fluid, a marbled (mosaic) appearance to the liver, and an enlarged, friable liver with a dilated gall bladder, splenomegaly, and congestion in gills, as shown in Figure 1F.
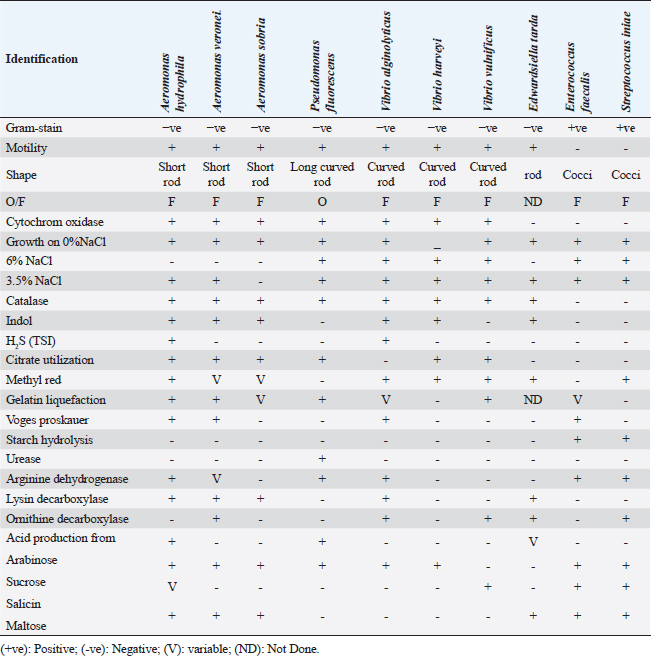
Fig. 1. Naturally infected O. niloticus showed body haemorrhages (Black arrow) and corneal opacity (Red arrow) with unilateral and bilateral exophthalmia (popeyes) (Yellow arrow) in (A–C), scale detachment (Blue arrow) in (D), fin degeneration and erosions (White arrow) in (E), while postmortem examination showed friable enlarged liver with marbling appearance (Yellow arrow), with splenomegaly (Red arrow) and congested gills (White arrow) in (F). Bacterial isolation and identificationThe naturally infected fish included 225 bacterial isolates, which were isolated. These isolates were phenotypically and biochemically characterized by the generally accepted technique published for bacterial isolation and identification. Aeromonas spp., Vibrio spp., Pseudomonas spp., Edwardsiella spp., Enterococcus spp., and Streptococcus spp. were among the isolates that were detected. All isolated bacterial strains have been identified phenotypically and biochemically, as shown in Table 2. Aeromonas hydrophila, sobria, and veronei were identified biochemically as separate species. The colonies of Aeromonas spp. on TSA medium looked glossy, spherical, creamy, and convex after 24 hours of incubation, but colonies on RS medium were light-yellow or deep creamy with the full edge. The isolated Aeromonas species were fermentative, catalase-producing, short rod, oxidase-positive, motile on semisolid medium, and Gram-negative. Table 2. The morphological and biochemical characteristics of isolated bacteria from naturally infected O. niloticus.
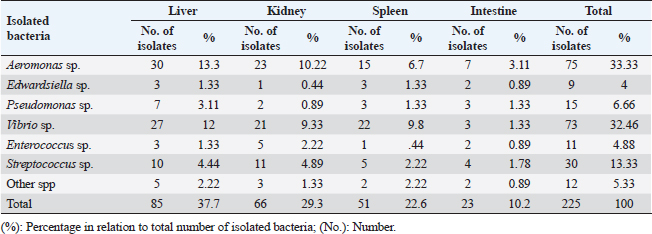
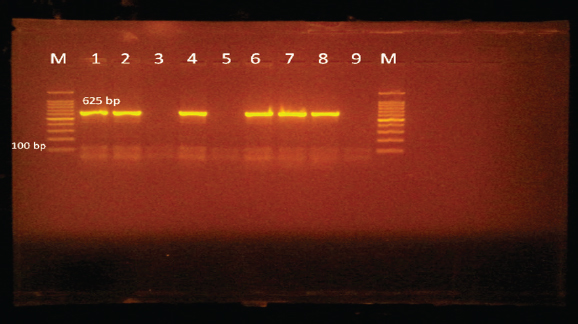
Edwardsiella tarda was further recognized biochemically as one of the Edwardsiella spp. All isolated Edwardsiella spp. emerged as greenish colonies with black centers over RS medium, which serves as a selective diagnostic media for E. tarda. The bacteria were determined to be motile, Gram-negative, short rod-shaped, and oxidase-negative organisms. In addition, Pseudomonas spp. colonization on TSA media displayed spindle form and diffusible dim yellow-green fluorescent pigments after 24 hours of incubation, whereas greenish colonies were noticed on the RS medium. On Pseudomonas-F-agar medium, however, it showed colonies that were 2–3 mm in diameter, yellowish green in color, and fluoresced after 48 hours of incubation. Pseudomona fluorescens was identified biochemically as Pseudomonas spp. All isolated Pseudomonas species were oxidase-positive, motile, Gram-negative, long, curved rods. However, additional biochemical identification was done for the Vibrio spp.; Vibrio harveyi, Vibrio alginolyticus, and Vibrio vulnificus. Bent rods, fermentative, Gram-negative, and positive for methyl red, catalase, and oxidase were all characteristics of the isolated Vibrio species. Vibrio vulnificus generated green colonies on TCBS media, whereas V. alginolyticus and V. harveyi generated yellow colonies. Colonies of Enterococcus sp. emerged on Streptococcus selective agar as extremely small, spherical, white colonies with complete borders or creamy rounded, big colonies (2–3 mm) with dew drop-like characteristics. An Enterococci displayed dark brown and black colonies on BBLTM EA. According to staining with Gram stain, Enterococcus faecalis is Gram-positive and is clustered in couples and occasionally short chains. Enterococcus faecalis is nonmotile bacteria and is negative for catalase, oxidase, and lysine decarboxylase. Biochemical testing identified S. iniae as one of the species of Streptococcus. It is a nonmotile, Gram-positive cocci that is typically organized in chains and is oxidase and catalase negative. Colonies typically have a greyish to pale color and shine over Edwards agar media. On blood agar, complete hemolysis was present in small, transparent, or pigmented white colonies up to 2 mm in diameter. Incidence of bacterial isolates in different organs and tissuesAccording to Table 2, 225 bacterial isolates were discovered in the liver, kidney, spleen, and intestine of naturally ill fish. The total amount of isolated bacteria found in various tissues (liver, kidney, spleen, and intestine) revealed six species; Aeromonas spp. (33.33%), Edwardsiella spp (4%), Pseudomonas spp (6.66%), Vibrio spp. (32.46%), Enterococcus spp. (4.88%), and Streptococcus spp. (13.33%). There were numerous instances of mixed infections in the same fish. Aeromonas spp. (75 isolates), Vibrio spp. (73 isolates), and Streptococcus spp. (30 isolates) were the three most common bacterial species. Prevalent bacterial isolates were chosen at random and tested by PCR. According to Table 3, the liver had the greatest percentage of isolated bacteria (37.7%), followed by the kidney (29.3%), spleen (22.6%), and the intestine (10.2%). In addition, the kidney, liver, and spleen yielded the bulk of the isolates of Aeromonas spp., Vibrio spp., and Streptococcus spp., while the gut had the lowest prevalence rates. Molecular identificationTo confirm the biochemically identified species, conventional PCR was used for molecular characterization. First, for the seven Aeromonas isolates chosen, Aeromonas spp. isolates identified utilizing the 16S rRNA region unique for A. hydrophila produced an amplified product of 625 bp size for five isolates, whereas two isolates were negative (Fig. 2). Examining seven isolates of Vibrio species for the genus gene (16s rRNA gene) allowed researchers to assess the genotypic characteristics of the species. As shown in Figure 3, five out of the seven isolates showed positive outcomes for the 16s rRNA gene, which has a molecular weight of 663 bp. Table 3. Prevalence and distribution of isolated bacteria from examined lesion samples.
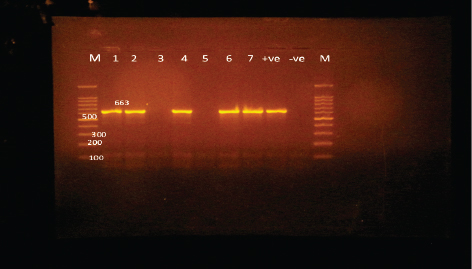
Fig. 2. Agarose gel electrophoresis of PCR products of 16S rRNA (625 bp) as species specific gene for confirmation of A. hydrophila isolated from O. niloticus. Lane M: 100–1,000 bp. DNA Ladder. Lane (1,2,4,6&7): positive A. hydrophila strains. Lane (3&5): negative A. hydrophila strains. Lane 8: positive control for A. hydrophila (at 625 bp). Lane 9: negative control for A. hydrophila.
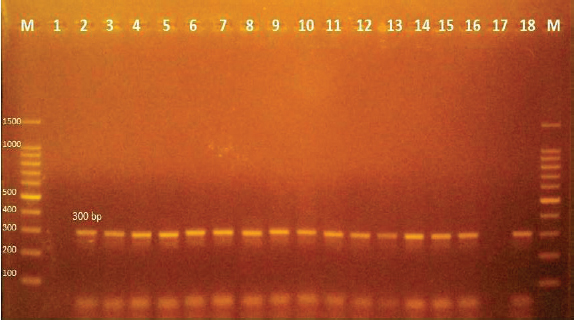
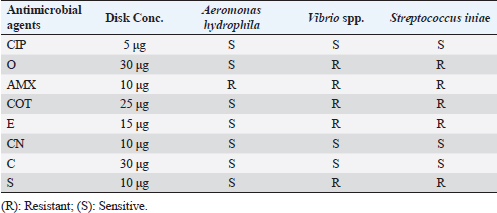
Fig. 3. Agarose gel electrophoresis of PCR products of 16S rRNA (663 bp) as species specific gene for confirmation of Vibro spp isolated from O. niloticus. Lane M: 100–1,000 bp. DNA Ladder. Lane (1,2,4,6&7): positive Vibro spp. Lane (3&5): negative Vibro spp. Lane +ve.: positive control for Vibro spp (at 663 bp.). Lane -ve .: Negative control or Vibro spp. Streptococcus isolates were identified using (Sin1, Sin2) specific genes for S. iniae, 16 Streptococcus isolates were tested and 15 were positive at 300 bp size, whereas 1 isolate was negative, as shown in Figure 4. Antimicrobial susceptibility assay of prevalent isolated bacteriaTo assess their susceptibility and the incidence of various types of antibiotic resistance among these isolates, the three common species isolates (A. hydrophila, Vibrio sp., and S. iniae) were tested against eight antimicrobial drugs in the current investigation (Table 4). The findings demonstrated that most A. hydrophila isolates were vulnerable to all antimicrobial substances, apart from AMX, against which they displayed resistance. While some strains of Vibrio and S. iniae were resistant to Oxytetracycline (O), AMX, CO-Trimethoprim-Sulfamethoxazole (COT), Erthyomycin (E), and S, certain strains of these bacteria were responsive to CIP, CN, and Chloramphenicol (C). All isolates showed high levels of resistance to AMX. However, for all isolated bacteria, CIP, CN, and C were the preferred medications.
Fig. 4. Agarose gel electrophoresis of PCR products of Sin1, Sin2 (300 bp) virulence genes for characterization of S. iniae. Lane M: 100-bp DNA ladder. Lane (2–16): positive S. iniae. Lane (1): negative S. iniae . Lane (17): negative control S. iniae. Lane (18): positive control S. iniae (300 bp). Table 4. Antibiotic sensitivity for A. hydrophila, Vibrio sp., and S. iniae.
DiscussionEven though Egypt’s aquaculture industry has had a remarkable development, it continues to face several difficulties due to disease outbreaks (Soliman and Yacout, 2016; ElSheshtawy et al., 2019). The most extensively cultivated species in Egypt is the Nile tilapia. Identical to other types of farmed fish, tilapia is sensitive to a variety of bacterial diseases that are common in freshwater environments, particularly in tough circumstances (Dong et al., 2017). Bacterial fish infections are a real threat to aquaculture (Bentzon-Tilia et al., 2016), resulting in outbreaks with large mortality rates (Sekar et al., 2008). The currently identified recovered pathogenic bacteria from farmed Nile tilapia constitute proof of the widespread geographic extent of pathogenic bacterial diseases. Current retrieved bacteria were recognized as Aeromonas spp. (Aeromonas sobria, A. hydrophila, and A. veronei), Edwardsiella spp. (E. tarda), Pseudomonas spp. (P. fluorescens.), Vibrio spp. (V. harveyi, V. alginolyticus, and V. vulnificus), Enterococcus spp. (Enterococcus faecalis), and Streptococcus spp. (Streptococcus iniae). The fish that were being examined exhibited extensive septicemic symptoms comparable to those mentioned by Austin and Austin (2007) and Matter et al. (2018). Such symptoms may be caused by the pathogenic enteric bacteria’s virulence components. It is speculated that pathogenic bacteria prove to be more virulent than nonpathogenic bacteria because they possess virulence genes. Both nonpathogenic and pathogenic bacteria encompass virulence genes, but only pathogenic bacteria can employ such genes (Moustafa et al., 2016). The anorexia and frayed and sloughed fins of the diseased fish may have contributed to their lethargy and sluggish movement, which ultimately affected their ability to swim and perform other bodily functions. The source of the hemorrhages throughout the body could be the hemolysin produced by the Aeromonas retrieved bacteria, which hemolyzes red blood cells (RBCs), in addition to the elastase enzyme, which significantly contributes to vascular injury because the blood arteries are primarily made up of elastic and collagenous fibers. The clinical symptoms seen in this study almost the same as those identified by previously (Ahmed and Shoreit, 2001; El-Ashram, 2002; Pretto-Giordano et al., 2010; Yardimci and Aydin, 2011; Noor El-Deen et al., 2014; Soliman et al., 2014; Othman et al., 2015; Dar et al., 2016; Omar et al., 2016; Abd El-Tawab et al., 2017; El-Barbary, 2017; Laith et al., 2017; Osman et al., 2017; Pech et al., 2017; Saleh et al., 2017; Amrullah et al., 2018; Khalil and Emeash, 2018; Hamouda et al., 2019; Hassan et al., 2020). Exophthalmia in the ongoing research may be a result of the S. iniae cytotoxic effects on the fish cells or fluid accumulation in the orbits behind the eyeballs (Locke et al., 2007). It is possible that the virulence factor directly causes cytotoxicity to the fish cells, loss of scales, and detachment as well as fins erosion. The virulence factors that S. iniae produce, which cause the death of blood cells and result in hemorrhage in many parts of the body, may also be responsible for the hemorrhagic patches over the mouth, operculum, and pectoral fins. S. iniae virulence factors may be responsible for eye cloudiness because they make it easier for Streptococcus to invade fish eyes, which causes cloudiness from bacterial proliferation (Moustafa et al., 2021). The postmortem inspection of the body in the current study revealed an accumulation of sanguineous ascitic fluid within the abdominal cavity, a mosaic-like pattern on the liver, and an enlarged, friable liver with a dilated gall bladder, splenomegaly, and clogged gills. The postmortem modifications could be caused by the identified septicemic pathogens and their virulence genes. The postmortem outcomes presented in this study are highly comparable to those investigated by a wide number of investigators in earlier publications (Pritchard and Lin, 1993; Ahmed and Shoreit, 2001; Khafagy et al., 2009; Musa et al., 2009; Umesha et al., 2011; Soliman et al., 2014; Dahdouh et al., 2016; Moustafa et al., 2016, Omar et al., 2016, 2017; Ortega et al., 2017; Osman et al., 2017; Saleh et al., 2017; Ahmed et al., 2018; El-Gamal et al., 2018; Hardi et al., 2018; Hamouda et al., 2019; Mansour et al., 2019; Hassan et al., 2020; Gobarah et al., 2021; Moustafa et al., 2021). In the current study, the assumed identification of the retrieved bacteria was conducted phenotypically through colony morphology on a selective differential medium for every suspected isolate; RS agar for Aeromonas spp. (Sarkar et al., 2013)., Pseudomonas sp. and Edwardsiella sp. (Buller, 2004; Acharya et al., 2007; Das et al., 2014); Pseudomonas-F-agar base medium for P. fluorescens (Omar et al., 2017), TCBS for Vibrio spp. (Gobarah et al., 2021); blood agar, Edwards Medium (modified), and Streptococcus selective agar for Streptococcus spp. (Saleh et al., 2017) and BBLTM EA for the isolation of Enterococci bacteria (Hamouda et al., 2019). Moreover, biochemical analyses seemed to be a useful technique for classifying and differentiating bacterial fish pathogens (Austin et al., 1997; Sarkar et al., 2012). Gram staining, colonies’ shape and biochemical activity are remarkably similar to those of the bacteria previously reported by a large number of prior authors (Ahmed and Shoreit, 2001; Masbouba, 2004; Khafagy et al., 2009; Noga, 2010; Soliman et al., 2014; Dar et al., 2016; Moustafa et al., 2016; Omar et al., 2016; Abd El-Kader and Mousa-Balabel, 2017; Abd El-Tawab et al., 2017; El-Barbary, 2017; Laith et al., 2017; Omar et al., 2017; Osman et al., 2017; Rahmatullah et al., 2017; Saleh et al., 2017; El-Gamal et al., 2018; Hardi et al., 2018; Ortega et al., 2018; Hamouda et al., 2019; Gobarah et al., 2021; Moustafa et al., 2020). Microbiological analysis of the affected fish showed the isolation of Gram-negative bacteria, with Aeromonas spp. having the highest prevalence (33.33%). These results may wholly or partially diverge from those noted by several authors and this may be referred to the abiotic and biotic circumstances of the habitats where the experiments were conducted. The current findings are consistent with some research that claimed Vibrio spp., Aeromonas spp., and Pseudomonas spp. were accountable for the fatal fish outbreak (Daskalov, 2006; Najiah et al., 2012; Youssuf et al., 2020). These findings are also consistent with those made available by El-Gamal et al. (2018), who found that the retrieved bacteria from naturally affected Nile tilapia from various fish farms in Kafr El-Sheikh Governorate, Egypt, were 26% Aeromonas spp., 23.3% Pseudomonas spp. 36.6% of mixed infections are the cause of ulcerative syndrome. This proved that the outbreak was predominantly caused by the same genera as before. To prevent the continued decline of fish farming, these strains must still be controlled. According to a previous study by Hardi et al. (2018) conducted in Indonesia, Gram-negative bacteria are the most often found bacteria reported in cultured tilapia. It has been discovered that tilapia and catfish have seven different bacterial genera: Staphylococcus sp., Streptococcus sp., Pseudomonas sp., Aeromonas sp., Enterobacter sp., Listeria sp., and Neisseria sp. The three most prevalent pathogenic bacteria in the current study were Aeromonas spp., members of the Vibrio genus, and Streptococcus spp. In fish, Aeromonas spp. produce motile Aeromonas septicemia and epizootic ulcerative syndrome, according to Austin and Austin (2012). Aeromonas hydrophila strains that were extremely virulent were found in many cases (Nielsen et al., 2001; Hemstreet, 2010). Aeromonas spp. (A. hydrophila), which made up around 33.33% of the infected fish farms, was followed by Vibrio spp. (32.46%), and Streptococcus spp. (13.33%). The same results from the same study region have been obtained (Joh et al., 2013; Walakira et al., 2014). Similar outcomes have been reported by other researchers (Newaj-Fyzul et al., 2008; Ribeiro et al., 2010), and this may be because Aeromonas species are common in the aquatic environment, the production systems utilized are comparable, and the fish species employed in the various trials. The majority of bacterial pathogens found in fish farms are common aquatic dwellers and do not simply arise as a result of an infectious agent being exposed to a host (Wedekind et al., 2010). The identified bacterial disease outbreaks were frequently triggered by environmental modifications (stressors), including O. niloticus overwintering, rapid temperature changes, inappropriate handling, injuries sustained during transit, and induced spawning in April and May every year. Fish’s homeostatic mechanism is susceptible to environmental stressors, which can lower the fish’s resistance to harmful organisms and cause illness outbreaks (Small and Bilodeau, 2005). Because of this, fish raised in aquaculture are more susceptible to a range of diseases (El-Sayed, 2006). Methicillin-resistant Staphylococcus aureus and A. sobria are two rare bacterial fish diseases that can infect tilapia, but because the aquaculture environment is in close contact with human and/or animal waste, these bacteria thrive there (Soliman et al., 2014; Dar et al., 2016). In terms of water temperature, the elevated warm temperature present in some fish farms may have a substantial effect and fish are becoming more vulnerable to bacterial diseases. The start and gravity of fish disease caused by Enterobacteriaceae-related bacterial pathogens were predominantly influenced by external circumstances such as contaminated water, high temperature, and excessive amounts of organic matter (Zheng et al., 2004). The stress of high water temperature promotes different virulent factors’ expression, speed up the spread of infection, increasing disease prevalence and the size of epidemics (Karvonen et al., 2010; Rodkhum et al., 2011; Kayansamruaj et al., 2014). According to the current investigation’s findings, the liver (37.7%), kidney (29.3%), spleen (22.6%), and intestine (10.2%) were the organs where isolated bacteria were found most frequently. These results align with earlier studies by Eissa et al. (2010), who clarified that Pseudomonas sp. was most prevalent in the liver (35%) and kidney (30%) of Nile tilapia, followed by the spleen (21.2%) and gills (13.7%), and El-Sayed (2019), where he revealed that naturally infected O. niloticus showed an elevated frequency of V. alginolyticus in the liver (38.5%), kidney (29.2%), spleen (23%), and heart (9.2%). In the current research, the liver, kidney, and spleen were the primary sources. In contrast, Mahmoud et al. (2016) clarified that the liver, spleen, and kidney were the primary locations of the largest distribution of bacterial isolates. According to Elsayed et al. (2018), the kidney and liver were used to isolate bacteria at rates of 37.4% and 36.3%, respectively. The spleen (15.9%) and gills (9.85%) were next in line. Furthermore, according to Eissa et al. (2016), the prevalence rates of A. sobria isolates were highest in the kidneys (25.3%), liver (23.0%), spleen (19.8%), and intestines (15.0%), while gills and skin lesions had the lowest frequency rates at 10.3% and 6.35%, respectively. Conventional morphological techniques are difficult and long-lasting, and they postpone the execution of control strategies, which results in significant financial losses. Furthermore, many pathogens induce comparable clinical signs in diseased fish and have similar morphological characteristics. As a result, molecular recognition approaches are used (Bader et al., 2003; Altinok, 2011; Tsai et al., 2012). The PCR has shown to be a more precise and efficient way to detect bacterial infections. The highly conserved 16S ribosomal RNA gene, which is found in bacteria, is essential for gene coding. In bacterial phylogenetic studies, it is recognized as a characteristic marker for distinguishing across species (Nagpal et al., 1998). In the present investigation, A. hydrophila was identified using the 16S rRNA gene which gave positive results at a molecular weight of 625 bp size. The outcomes correspond with those of earlier research (Gordon et al., 2007; Abd El-Tawab et al., 2019); however, in contrast to Omar et al. (2016), where A. hydrophila was identified at a molecular weight 953 bp size and Hamouda et al. (2019), where A. hydrophila was identified at a molecular weight of 356 bp size. PCR identified Vibrio species, validating the genus (16S rRNA) in all of the isolates at 663 bp. This outcome was consistent with that reported by Gobarah et al. (2021). Moreover, it is interesting to note that the sin1 and sin2 virulence genes discovered in the current investigation were exclusive to pathogenic S. iniae. The results obtained agreed with what was mentioned by Baiano and Barnes (2009); Moustafa et al. (2021). Research on the susceptibility of fish pathogens to antibiotics is crucial in the development of novel chemotherapeutic medications for bacterial infection treatment in specific populations of cultivated fish. To ascertain the sensitivity and frequency of various antibiotic resistance between the bacterial isolates during the summer mortality of farmed Nile tilapia, eight antimicrobial agents were used. Sensitivity testing clarified that most of the strains tested were susceptible to CIP, CN, and C. Therefore, these antibiotics are effective against these bacteria and can be used to treat affected fish (Drug of choice). However, all isolates displayed high levels of resistance to AMX, indicating that these antibiotics should not be used to treat fish with pathogenic bacterial infections. The outcomes are remarkably consistent with those of Dahdouh et al. (2016), who discovered that freshwater fish isolate A. hydrophila was responsive to ofloxacin, enrofloxacin, and CN but resistant to erythromycin. According to El-Barbary (2017), A. hydrophila was resistant to Erythromycin and Penicillin but susceptible to Norfloxacin, CIP, Lomefloxacin, Gatifloxacin, Kanamycin, and Gentamycin. According to research by Hamouda et al. (2019), A. hydrophila isolates were sensitive to the following drugs (in decreasing order): CN, CIP, Amikacin, Nalidixic acid, Cefotaxim, Cephalothin, Kanamycin, Sulfamethoxazole, and Ampicillin and completely resistant to Penicillin and Erythromycin. Similarly to this, El-Barbary and Hal (2016) showed that P. fluorescens, A. hydrophila, P. putida, and Klebsiella-infected fish may be treated with CIP, Norfloxacin, and Gentamycin. While most isolates of A. hydrophila in our investigation displayed resistance to O, Ahmed and Shoreit (2001) demonstrated that Aeromonas sp. retrieved from O. niloticus from Aswan fish hatcheries was extremely sensitive to the antibiotic. This shows that drug resistance to O in this region has appeared as a consequence of aquaculture’s foolish use of antibiotics. In addition, the current finding regarding Vibrio spp. revealed widespread antibiotic resistance found in Vibrio spp. retrieved from fish. There was a prevalent resistance to erythromycin. The outcomes are coincided with previous reports (Kitiyodom et al., 2010; Gobarah et al., 2023). This may be attributed to that erythromycin is naturally produced and subsequently scattered around in the environment, this is not surprising (Rosser and Young, 1999; Bani et al., 2007). However, the current investigation disagrees with Gobarah et al. (2023), where Vibrio spp. revealed sensitivity to Gentamycin. Streptococcus iniae isolates, on the other hand, revealed that every isolate examined was susceptible to CIP, CN, and C and resistant to O, AMX, Erythromycin, and S. The current investigation agrees with Younes et al. (2019) where S. iniae was resistant to AMX but, at the same time disagrees with the same author as S. iniae is sensitive to O in their study. ConclusionAccording to the current investigations, A. hydrophila, Vibrio sp., and S. iniae are the most threatening prevalent bacterial pathogens that have been isolated during mass-farmed tilapia summer mortality. This is in line with the hypothesis that high water temperatures may enhance the risk of these bacterial infections, resulting in mass tilapia mortality and significant economic losses. To raise healthy fish, it is therefore essential to maintain a favorable fish culture, environment, and husbandry techniques so that enhancing and improving the tilapia-rearing environment and reducing the pathogen’s virulence. The majority of the tested strains were susceptible to CIP, CN, and C; but, displayed high levels of resistance to AMX. AcknowledgmentThe Central Diagnostic Research Laboratory, Fac. Vet. Med., Kafrelsheikh Univ. is gratefully acknowledged by the authors for its assistance with the PCR technology. Fish Diseases and Management Department, Fac. Vet. Med., Kafrelsheikh University is acknowledged by the authors for providing the bacterial Reference strains, necessary resources, and assistance during the research procedure. Conflict of interestThe authors have not disclosed any conflicting interests. FundingThis work was entirely supported by the authors’ contributions; there was no outside financial source. Author contributionsThe work’s purpose and design were contributed by all of the authors. The study’s conception and design were created by Fawzy Magouz and Eman Moustafa. In addition, Eman Moustafa, Amira Omar, Etab Abo-Remela, and Passant Barakaat prepared the material and handled data collection and analysis. Marwa Halawa conducted research on antibiotic sensitivity and the PCR technique. The manuscript’s first draft was written by Eman Moustafa, Amira Omar, Etab Abo-Remela, Marwa Halawa, and Passant Barakaat. Each author provided feedback on an earlier version of the manuscript. Eman Moustafa then reviewed and corrected the final manuscript. The final version of the article was read and endorsed by every involved author. Data availabilityAll of the information necessary to contribute to the conclusions of this investigation is included in the current paper. The corresponding author will supply any additional information required upon reasonable request. ReferencesAbbas, E.M., Soliman, T., El-Magd, M., Farrag, M., Ismail, R.F. and Kato, M. 2017. Phylogeny and DNA barcoding of the family Sparidae inferred from mitochondrial DNA of the Egyptian waters. J. Fish. Aquat. Sci. 12, 73–81. Abd El-Kader, M. and Mousa-Balabel, T. 2017. Isolation and molecular characterization of some bacteria implicated in the seasonal summer mortalities of farm-raised Oreochromis niloticus at Kafr El-Sheikh and Dakahlia governorates. Alex. J. Vet. Sci. 53(2), 107–113. Abdelsalam, M., Asheg, A. and Eissa, A. 2013. Streptococcus dysgalactiae: an emerging pathogen of fishes and mammals. Int. J. Vet. Sci. Med. 1, 1–6. Abdelsalam, M., Chen, S. and Yoshida, T. 2010. Phenotypic and genetic characterizations of Streptococcus dysgalactiae strains isolated from fish collected in Japan and other Asian countries. FEMS. Microbiol. Lett. 302, 32–38. Abd El-Tawab, A.A., Maarouf, A.A., El-Hofy, F.I. and El-Mougy, E.A. 2017. Detection of some virulence genes in A. hydrophila and A. caviae isolated from fresh water fishes at Qalubia Governorate. Benha. Vet. Med. J. 33(2), 489–503. Abd El-Tawab, A.A., Maarouf, A.A.A., El-Hofy, F.I., Salim, A.O. and Elsayed, A.A.M. 2019. Phenotypic and molecular detection of Aeromonas and Pseudomonas species isolated from fish with special reference to their virulence factors. Nat. Sci. 17(12), 194–205. Acar, U., Kesbic, O.S., Yılmaz, S., Gultepe, N. and Turker, A. 2015. Evaluation of the effects of essential oil extracted from sweet orange peel (Citrus sinensis) on growth rate of tilapia (Oreochromis mossambicus) and possible disease resistance against Streptococcus iniae. Aquacult 473, 282–286. Acharya, M., Maiti, N.K., Mohanty, S., Mishra, P. and Samanta, M. 2007. Genotyping of Edwardsiella tarda isolated from freshwater fish culture system. Comp. Immun. Microb. Inf. Dis. 30, 33–40. Adelekea, B., Robertson-Anderssona, D., Moodleya, G. and Taylorb, S. 2021. Aquaculture in Africa: a comparative review of Egypt, Nigeria, and Uganda Vis-A-Vis South Africa. Rev. Fish. Sci. Aquac. 29(2), 167–197. Ahmed, H.A., Mohamed, M.E.M., Rezk, M.M., Gharieb, R.M.A. and Abdel-Maksoud, S.H. A. 2018. Aeromonas hydrophila in fish and humans; prevalence, virulo-typing and antimicrobial resistance. Slov. Vet. Res. 55(Suppl 20), 113–124. Ahmed, S.H.M. and Shoreit A.M. 2001. Bacterial hemorrhagic septicemia in Oreochromis niloticus at Aswan fish hatcheries. Assiut. Vet. Med. J. 45(89), 190–206. Algammal, M.A., Mohamed, M.F., Tawfiek, B.A., Hozzein, W.N., El Kazzaz, W.M. and Mabrok, M. 2020. Molecular typing, antibiogram and PCR-RFLP based detection of Aeromonas hydrophila complex isolated from Oreochromis niloticus. Pathogens 9, 238. Ali, M.M., Ali, M.L., Proshad, R., Islam, S., Rahman, Z., Tusher, T.R., Kormoker, T. and Abdullah, M.A. 2020. Heavy metal concentrations in commercially valuable fishes with health hazard inference from Karnaphuli River, Bangladesh. Hum. Ecol. Risk. Assess. 26, 10. Altinok, I. 2011. Multiplex PCR assay for detection of four major bacterial pathogens causing rainbow trout disease. Dis. Aquat. Organ. 93, 199–206. Amrullah, B.I., Jaya, A. and Wahida, A. 2018. Streptococcus agalactiae whole cell bacteria toxin protein in Nile tilapia Oreochromis niloticus. AACL. Bioflux. 11(2), 460–468. Austin, B., Alsina, M., Austin, D.S., Blanch, A.R., Grimont, F. and Grimont, P.A.D. 1997. A comparison of methods for the typing of fish pathogenic Vibrio spp. System. Appl. Microb. 20(1), 89–101. Austin, B. and Austin, D.A. 2007. Bacterial fish pathogens, disease of farmed and wild fish, 4th ed. Godalming, UK: Springer Praxis. Austin, B. and Austin, D.A. 2012. Aeromonadaceae representatives (motile aeromonads). Bacterial fish pathogens. Berlin, Germany: Springer, pp: 119–146. Bader, J.A., Shoemaker, C.A. and Klesius, P.H. 2003. Rapid detection of columnaris disease in channel catfish (Ictalurus punctatus) with a new species specific 16S rRNA gene-based PCR primer for Flavobacterium columnare. J. Microbiol. Meth. 52, 209–220. Baiano, J.C.F. and Barnes, A.C. 2009. Towards control of Streptococcus iniae. Emerg. Inf. Dis. 15(12), 1891–1896. Bani, S., Mastromarino, P.N., Ceccarelli, D., Le Van, A., Salvia, A.M., Ngo Viet, Q.T., Hai, D.H., Bacciu, D., Cappuccinelli, P. and Colombo, M.M. 2007. Molecular characterization of ICEVchVie0 and its disappearance in Vibrio cholerae O1 strains isolated in 2003 in Vietnam. FEMS. Microbiol. Lett. 266, 42–48. Bentzon-Tilia, M., Sonnenschein, E.C. and Gram, L. 2016. Monitoring and managing microbes in aquaculture –towards a sustainable industry. Microb. Biotech. 9(5), 576–584. Bergey, D.H. 1994. Bergey’s manual of determinative bacteriology, 9th ed. Eds., Buchaman, R.E. and Gibbons, N.E. Baltimore, MD: Williams and Wilkins. Bulfon, C., Volpatti, D. and Galeotti, M. 2015. Current research on the use of plant- derived products in farmed fish. Aquac. Res. 46, 513–551. Buller, N.B. 2004. Bacteria from fish and other aquatic animals: a practical identification manual. Oxford, UK: CABI Publishing. Available via https://www.cabi.org/bookshop/book/9781845938055/ Chen, C., Chao, C. and Bowser, P.R. 2007. Comparative histopathology of Streptococcus iniae and Streptococcus agalactiae-infected tilapia. Bull. Eur. Assoc. Fish. Pathol. 27, 2–29. CLSI. 2016. Performance standards for antimicrobial susceptibility testing, 26th ed. Wayne, PA: CLSI supplement M100S.Cruickshank, R., Duguid, J.P., Marmion, B.P. and Swain, R.H. 1975. Medical microbiology. The practical of microbiology, 12th ed. Edinburgh, London, UK; and New York, NY: Chucohill Livingstone. Dahdouh, B., Basha, O., Khalil, S. and Tanekhy, M. 2016. Molecular characterization, antimicrobial susceptibility and salt tolerance of Aeromonas hydrophila from fresh, brackish and marine fishes. Alex. J. Vet. Sci. 48(2), 46–53. Dar, G.H., Kamili, A.K., Chishti, M.Z., Dar, S.A., Tantry, T.A. and Ahmad, F. 2016. Characterization of Aeromonas sobria isolated from fish Rohu (Labeo rohita) collected from polluted pond. J. Bacteriol. Parasitol. 7, 273. Das, B.K., Sahu, I., Kumari, S., Sadique, M. and Nayak, K.K. 2014. Phenotypic and whole cell protein profiling of Edwardsiella tarda strains isolated from freshwater fishes. Int. J. Curr. Microb. Appl. Sci. 3(1), 235–247. Daskalov, H. 2006. The importance of Aeromonas hydrophila in food safety. Food. Con. 17(6), 474–483. Dickson, M., Nasr-Allah, A., Kenawy, D., Fathi, M., El-Naggar, G. and Ibrahim, N. 2016. Improving employment and income through development of Egypt’s aquaculture sector (IEIDEAS) project. WorldFish. Dong, H., Techatanakitarnan, C., Jindakittikul, P., Thaiprayoon, A., Taengphu, S., Charoensapsri, W. and Senapin, S. 2017. Aeromonas jandaei and Aeromonas veronii caused disease and mortality in Nile tilapia, Oreochromis niloticus (L.). J. Fish. Dis. 40(10), 1395–1403. Eissa, A.E. 2016. Clinical and laboratory manual of fish diseases. Saarbrücken, Germany: LAP LAMBERT Academic Publishing. Eissa, I.A.M., Derwa, H.I., El-Lamei, M., Desuki, A., Zaki, M.S. and El-Sheshtawy, H. 2013. Iron in water and some marine fishes in relation to vibriosis at Lake Temsah. Life. Sci. J. 10(3), 2520–2528. Eissa, N.M.E., El-Ghiet, E.A., Shaheen, A.A. and Abbass, A. 2010. Characterization of Pseudomonas species isolated from tilapia “Oreochromis niloticus” in Qaroun and Wadi-El-Rayan lakes, Egypt. Glob. Vet. 5(2), 116–121. Eissa, I., El-Lamei, M., Ismail, T., Youssef, F. and Mansour, S. 2016. Advanced studies for diagnosis of aeromonas septicemia in Oreochromis niloticus. Suez. Canal. Vet. Med. J. 21(1), 221–233. Eissa, I.A.M., El-lamei, M., Sherif, M., Desuky, E., Zaki, M. and Bakry, M. 2015. Aeromonas veronii biovar sobria a causative agent of mass mortalities in cultured Nile tilapia in El-Sharkia governorate, Egypt. Life. Sci. J. 12(5), 90–97. El-Ashram, A.M. 2002. On Aeromonas hydrophila infection among cultured tilapias: a biological, histopathological and management study. Egypt. J. Aquat. Biol. Fish. 6(3), 181–202. El-Barbary, M.I. 2017. Serum biochemical and histopathological changes associated with Aeromonas hydrophila isolated from Oreochromis niloticus and Sparus aurata with multiple antibiotic resistance index. J. Biol. Sci. 17, 222–234. El-Barbary, M.I. and Hal, A.M. 2016. Isolation and molecular characterization of some bacterial pathogens in El-Serw fish farm, Egypt. Egypt. J. Aquat. Biol. Fish. 20(4), 115–127. El-Gamal, A.M., El-Gohary, M.S. and Gaafar, A.Y. 2018. Detection and molecular characterization of some bacteria causing skin ulceration in cultured Nile tilapia (Oreochromis niloticus) in Kafr El-Sheikh governorate. Int. J. Zool. Res. 14, 14–20. El-Sayed, A.M. 2006. Tilapia culture. Oxfordshire, UK: CABI Publishing, CAB International. https://doi.org/10.1079/9780851990149.0000El-Sayed, A.F.M. 2019. Tilapia culture. Cambridge, MA: Academic Press. Elsayed, M., Essawy, M., Shabana, I., Abou El-Atta, M. and EL-Banna, N. 2018. Studies on bacterial pathogens in some marine fishes in EL-Mansoura, Egypt. Amer. J. Agric. Biol. Sci. 13(1), 9–15. Elsheshtawy, A., Yehia, N., Elkemary, M. and Soliman, H. 2019. Investigation of Nile tilapia summer mortality in Kafr El-Sheikh Governorate, Egypt. Genet. Aquat. Org. 3(1), 17–25. Eltholth, M., Fornace, K., Grace, D., Rushton, J. and Häsler, B. 2015. Characterisation of production, marketing and consumption patterns of farmed tilapia in the Nile Delta of Egypt. Food. Policy. 51, 131–143. Fang, U.X., Stefan, B.H.G., Eaton, J.G., McCormick, J.H. and Alama, S.R. 2004. Simulation of thermal/dissolved oxygen habitat for fishes in lakes under different climate scenarios Part 1. Cool-water fish in the contiguous. Ecol. Model. 172, 13–37. Fathi, M., Dickson, C., Dickson, M., Leschen, W., Baily, J., Muir, F., Ulrich, K. and Weidmann, M. 2017. Identification of Tilapia Lake virus in Egypt in Nile tilapia affected by ‘summer mortality’ syndrome. Aquaculture 473, 430–432. Gobarah, D.E.A., Helmy, M., Mahfouz, N.B., Fahmy, H.A., Abou Zeid, M.A.M. and Moustafa, E.M. 2021. Phenotypic and molecular characterization of Vibrio species isolated from fish markets in Egypt. J. Hell. Vet. Med. Soc. 72(2), 2817–2824. Gobarah, D.E.A., Helmy, M., Mahfouz, N.B., Fahmy, H.A., Abou Zeid, M.A.M. and Moustafa, E.M. 2023. Molecular characterization of antimicrobial resistance of Vibrio species isolated from fish in Egypt. J. Hell. Vet. Med. Soc. 74(1), 5101–5110. Gordon, L., Giraud, E., Ganiẽre, G.P., Armand, F., Bouju-Albert, A., de la Cotte, N., Mangion, C. and Le Bri, H. 2007. Antimicrobial resistance survey in a river receiving effluents from freshwater fish farms. J. Appl. Microbiol. 102, 1167–1176. Haldar, S., Majumdar, S., Chakravorty, S., Tyagi, J., Bhalla, M. and Sen, M. 2005. Detection of acid-fast bacilli in postlysis debris of clinical specimens improves the reliability of PCR. J. Clin. Microbiol. 43, 3580–3581. Hamouda, A.H., Moustafa, E.M. and Zayed, M.M. 2019. Overview on the most prevailing bacterial diseases infecting Oreochromis niloticus at Aswan fish hatchery, Egypt. Adv. Anim. Vet. Sci. 7(11), 950–961. Hardi, E.H., Nugroho, R.A., Saptiani, G., Sarinah, R., Agriandini, M. and Mawardi, M. 2018. Identification of potentially pathogenic bacteria from tilapia (Oreochromis niloticus) and channel catfish (Clarias batrachus) culture in Samarinda, East Kalimantan, Indonesia. Biodiversitas 19(2), 480–488. Harvell, C.D., Kim, K., Burkholder, J.M., Colwell, R.R., Epstein, P.R., Grimes, D.J., Hofmann, E.E., Lipp, E.K., Osterhaus, A., Overstreet, R.M., Porter, J.W., Smith, G.W. and Vasta, G.R. 1999. Marine ecology—emerging marine diseases—climate links and anthropogenic factors. Science 285, 1505–1510. Hassan, S., Abdel-Rahman, M., Mansour, E.S. and Monir, W. 2020. Prevalence and antibiotic susceptibility of bacterial pathogens implicating the mortality of cultured Nile tilapia, Oreochromis niloticus. Egypt. J. Aquacul. 10(1), 23–43. Hassan, M.A., Noureldin, E., Mahmoud, M.A. and Fita, N.A. 2017. Molecular identification and epizootiology of Aeromonas veronii infection among farmed Oreochromis niloticus in Eastern Province, KSA. Egypt. J. Aquat. Res. 43(2), 161–167. Hemstreet, B. 2010. An update on Aeromonas hydrophila from a fish health specialist for summer. Catfish. J. 24, 4. Hussain, S.M., Javed, M., Javid, A., Javid, T. and Hussain, N. 2011. Growth responses of Catla catla, Labeo rohita and Cirrhina mrigala during chronic exposure of iron. Pak. J. Agric. Sci. 48(3), 225–230. Joh, S.J., Ahn, E.H., Lee, H.J., Shin, G.W., Kwon, J.H. and Park, C.G. 2013. Bacterial pathogens and flora isolated from farm-cultured eels (Anguilla japonica) and their environmental waters in Korean eel farms. Vet. Microb. 163(1-2), 190–195. Karvonen, A., Rintamäki, P., Jokela, J. and E. Valtonen, T. 2010. Increasing water temperature and disease risks in aquatic systems: climate change increases the risk of some, but not all, diseases. Int. J. Parasit. 40, 1483–1488. Kayansamruaj, P., Pirarat, N., Hirono, I. and Rodkhum, C. 2014. Increasing of temperature induces pathogenicity of Streptococcus agalactiae and the up-regulation of inflammatory related genes in infected Nile tilapia (Oreochromis niloticus). Vet. Microbiol. 172, 265–271. Khafagy, A., Eid, H., Abo, El-Atta, M. and Abd El-Fattah, L. 2009. Isolation of Enterococcus faecalis from tilapia in Lake Temsah in Ismailia governorate. SCVMJ IVX(2), 45–54. Khalil, F. and Emeash, H. 2018. Behaviors and stereotypies of Nile tilapia (Oreochromis niloticus) in response to experimental infection with Aeromonas hydrophila. Aquat. Sci. Eng. 33(4), 124–130. Kitiyodom, S., Khemtong, S., Wongtavatchai, J. and Chuanchuen R. 2010. Characterization of antibiotic resistance in Vibrio spp. isolated from farmed marine shrimps (Penaeus monodon). FEMS. Microbiol. Ecol. 72(2), 219–227. Konemann, E., Allen, S., Janda, W., Schreckenberger, C. and Winn, W. 1997. Color atlas and textbook of diagnostic microbiology, 5th ed. Philadelphia, PA; New York, NY: Lippincott. Kreig, N. and Holt, J. 1984. Bergey’s manual of systemic bacteriology. Baltimore, MD: William and Wilkins, vol. 1. Laith, A.A., Ambak, M.A., Hassan, M., Sheriff, S.M., Nadirah, M., Draman, A.S., Wahab, W., Ibrahim, W.N.W., Aznan, A.S., Jabar, A. and Najiah, M. 2017. Molecular identification and histopathological study of natural Streptococcus agalactiae infection in hybrid tilapia (Oreochromis niloticus). Vet. World. 10(1), 101–111. Liamnimitr, P., Mon-on, N., Jaemwimon, P., Dachavichitlead, W. and Surachetpong, W. 2017. Detection of Streptococcus iniae and Streptococcus agalactiae in tilapia (Oreochromis sp.) in Thailand. Thai. J. Vet. Med. Suppl. 47, 285–286. Locke, J.B., Colvin, K.M., Varki, N., Vicknair, M.R., Nizet, V. and Buchanan, J.T. 2007. Streptococcus iniae β-hemolysin streptolysin S is a virulence factor in fish infection. Dis. Aquat. Org. 76, 17–26. MacFaddin, J.F. 2000. Biochemical tests for identification medical bacteria. Baltimore, MD: Warery Press Inc.Mahmoud, M.A., Abdelsalam, M., Mahdy, O.A., El Miniawy, H.M.F., Ahmed, Z.A.M., Osman, A.H., Mohamed, H.M.H., Khattab, A.M. and Ewiss, M.A.Z. 2016. Infectious bacterial pathogens, parasites and pathological correlations of sewage pollution as an important threat to farmed fishes in Egypt. Environ. Poll. 219, 939–948. Mansour, A., Mahfouz, N.B., Husien, M.M. and El-Magd, M.A. 2019. Molecular identification of Aeromonas hydrophila strains recovered from Kafrelsheikh fish farms. Slov. Vet. Res. 56(Suppl 22), 201–208. Markey, B.K., Leonard, F.C., Archambault, M., Cullinane, A. and Maguire, D. 2013. Clinical veterinary microbiology, 2nd ed. London, UK: MOSBY, Elsevier Ltd. Masbouba, I.M.M. 2004. Studies on Pseudomonas infection in fish in Kafr El-Sheikh Province. MVSc. Thesis, Faculty of Veterinary Medicine, Tanta University, Tanta, Egypt. Matter, A.F., El-Asely, A.M., Shaheen, A.A., Abd El-Gawad, E.A., El-Abd, H. and Abbass, A.A. 2018. Phenotypic and molecular characterization of bacterial pathogens isolated from diseased freshwater fishes. Int. J. Fish. Aquat. Stud. 6(2), 34–41. Mohamed, H.M., Emeish, W.F., Braeuning, A. and Hammad, S. 2017. Detection of aflatoxin-producing fungi isolated from Nile tilapia and fish feed. EXCLI. J. 16, 1308–1318. ISSN 1611–2156. Moustafa, E.M., Dawood, M.A.O., Assar, D.H., Omar, A.A., Elbialy, Z.I., Farrag, F.A., Shukry, M. and Zayed, M.M. 2020. Modulatory effects of fenugreek seeds powder on the histopathology, oxidative status, and immune related gene expression in Nile tilapia (Oreochromis niloticus) infected with Aeromonas hydrophila. Aquaculture 515, 734589. Moustafa, E.M., Farrag, F.A., Dawood, M.A.O., Shahin, K., Hamza, A., Decamp, O., Mohamed, R., Elsabagh, M., Eltholth, M. and Omar, A.A. 2021. Efficacy of Bacillus probiotic mixture on the immunological responses and histopathological changes of Nile tilapia (Oreochromis niloticus, L) challenged with Streptococcus iniae. Aquac. Res. 52, 2205–2219. Moustafa, E.M., Omar, A.A. and Abdo, S.W. 2016. Insight into the virulence-related genes of Edwardsiella tarda isolated from cultured freshwater fish in Egypt. World. Vet. J. 6(3), 101–109. Musa, N., Wei, L.S., Musa, N., Hamdan, R.H., Leong, L.K., Wee, W., Amal, M.N., Kutty, B.M. and Abdullah, S.Z. 2009. Streptococcosis in red hybrid tilapia (Oreochromis niloticus) commercial farms in Malaysia. Aquac. Res. 40(5), 630–632. Nagpal, M.L., Fox, K.F. and Fox, A. 1998. Utility of 16S–23S rRNA spacer region methodology: how similar are interspace regions within a genome and between strains for closely related organisms. J. Microbiol. Methods. 33, 211–219. Najiah, M., Aqilah, N.I., Lee, K.L., Khairulbariyyah, Z., Mithun, S., Jalal, K.C.A., Shaharom-Harrison, F. and Nadirah, M. 2012. Massive mortality associated with Streptococcus agalactiae infection in cage-cultured red hybrid tilapia Oreochromis niloticus in Como River, Kenyir Lake, Malaysia. J. Biol. Sci. 12(8), 438–442. Newaj-Fyzul, A., Mutani, A., Ramsubhag, A. and Adesiyun, A. 2008. Prevalence of bacterial pathogens and their anti- microbial resistance in tilapia and their pond water in Trinidad. Zoon. Public. Health. 55(4), 206–213. Neyts, K., Huys, G., Uyttendaele, M., Swings, J. and Debevere, J. 2000. Incidence and identification of mesophilic Aeromonas spp. from retail foods. Lett. Appl. Microbiol. 31(5), 359–363. Nicholson, P., Mon-on, N., Jaemwimol, P., Tattiyapong, P. and Surachetpong, W. 2020. Coinfection of tilapia lake virus and Aeromonas hydrophila synergistically increased mortality and worsened the disease severity in tilapia (Oreochromis spp.). Aquaculture 520, 734746. Nicky, B.B. 2004. Bacteria from fish and other aquatic animals (a practical identification manual). Oxford, UK: CABI Publishing is a Division of CAB International, vol. 106, no. (85), pp: 83–116. Nielsen, M.E., Høi, L., Schmidt, A.S., Qian, D., Shimada, T., Shen, J.Y. and Larsen, J.L. 2001. Is Aeromonas hydrophila the dominant motile Aeromonas species that causes disease outbreaks in aquaculture production in the Zhejiang Province of China? Dis. Aquat. Org. 46(1), 23–29. Noga, E.J. 2010. Fish diseases, diagnosis and treatment, 2nd ed. Ames, IA: Iwa State University, Press. https://doi.org/10.1002/9781118786758 Noor El Deen, A.E., Dorgham, S.M., Hassan, A.H.M. and Hakim, A.S. 2014. Studies on Aeromonas hydrophila in cultured Oreochromis niloticus at Kafr El Sheikh Governorate, Egypt with reference to histopathological alterations in some vital organs. World. J. Fish. Marine. Sci. 6(3), 233–240. Omar, A.A., Moustafa, E.M., Abo-Remela, E.M. and Zayed, M.M. 2017. Prevalence, molecular characterization, pathogenecity and antimicrobial susceptibility of Pseudomonas fluorescens isolated from Oreochromis niloticus. Life. Sci. J. 14(8), 53–61. Omar, A.A., Moustafa, E.M. and Zayed, M.M. 2016. Identification and characterization of virulence- associated genes from pathogenic Aeromonas hydrophila strains. World’s. Vet. J. 6(4), 185–192. Ortega, Y.A., Barreiro, F.S., Castro, G.S., Huancaré, K.P., Manchego, A.S., Marco, A.A., Mayra, A.P., Wilson, G. and Sandoval, N.C. 2017. Beta-haemolytic streptococci in farmed Nile tilapia, Oreochromis niloticus, from Sullana-Piura, Peru. Rev. MVZ. Córdoba. 22(1), 5653–5665. Ortega, C.E.S.A.R., García, I., Irgang, R., Fajardo, R., Tapia-Cammas, D., Acosta, J. and Avendaño-Herrera, R. 2018. First identification and characterization of Streptococcus iniae obtained from tilapia (Oreochromis aureus) farmed in Mexico. J. Fish. Dis. 41(5), 773–782. Osman, K.M., Al-Maary, K.S., Mubarak, A.S., Dawoud, T.M., Moussa, I.M., Ibrahim, M.S., Hessain, A.M., Orabi, A. and Fawzy, N.M. 2017. Characterization and susceptibility of Streptococci and Enterococci isolated from Nile tilapia (Oreochromis niloticus) showing septicaemia in aquaculture and wild sites in Egypt. BMC. Vet. Res. 13, 357. Othman, F., Islam, M.S., Sharifah, E.N., Shahrom-Harrison, F. and Hassan, A. 2015. Biological control of streptococcal infection in Nile tilapia Oreochromis niloticus (Linnaeus, 1758) using filter-feeding bivalve mussel Pilsbryoconcha exilis (Lea, 1838). J. Appl. Ichthyol. 31, 724–728. Pech, G.H., Chavez, C.M.R. and Reynoso, F.L. 2017. Pathogenic bacteria in Oreochromis niloticus Var. Stirling tilapia culture. Fish. Aqua. J. 8, 197. Pretto-Giordano, L.G., Müller, E.E., Freitas, J.C.D. and Silva, V.G.D. 2010. Evaluation on the pathogenesis of Streptococcus agalactiae in Nile tilapia (Oreochromis niloticus). Braz. Arch. Biol. Tech. 53(1), 87–92. Pridgeon, J.W. and Klesius, P.H. 2012. Major bacterial diseases in aquaculture and their vaccine development. CAB. Rev. 7, 1–16. Pritchard, D.G. and Lin, B. 1993. Group B streptococcal neuraminidase is actually a hyaluronidase. Infect. Immun. 61, 3234–3239. Quinn, P.T., Markey, B.K., Carter, M.E., Donnelly, W.J and Leonard, F.C. 2002. Veterinary microbiology and microbial disease. Ames, IA: First Published Blackwell Science Company, Iowa State University Press. Rahmatullah, M., Ariff, M., Kahieshesfandiari, M., Daud, H.M., Zamri-Saad, M., Sabri, M.Y., Amal, M.N.A. and Ina-Salwany, M.Y. (2017). Isolation and pathogenicity of Streptococcus iniae in cultured red hybrid tilapia in Malaysia. J. Aquat. Anim. Health. 29, 208–213. Reda, R.M., Nasr, M.A.F., Ismail, T.A. and Moustafa, A. 2021. Immunological responses and the antioxidant status in African catfish (Clarias gariepinus) following replacement of dietary fish meal with plant protein. Animals 11, 1–19. Reynolds, J. (2011). Bacterial colony morphology. Dallas, TX: Richland College. Ribeiro, R.V., Reis, E.M.F., Reis, C.M.F., Freitas-Almeida, A.C. and Rodrigues, D.P. 2010. Incidence and antimicrobial resistance of entero-pathogens isolated from an integrated aquaculture system. Lett. Appl. Microbiol. 51(6), 611–618. Rodkhum, C., Kayansamruaj, P. and Pirarat, N. 2011. Effect of water temperature on susceptibility to Streptococcus agalactiae serotype in infection in Nile tilapia Oreochromis niloticus (L.). Thai. J. Vet. Med. 41, 309–314. Rosser, S.J. and Young, H.K. 1999. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J. Antimicrob. Chemoth. 44, 11–18. Saleh, H.A., Sabry, N.M., Abd Al-Razik, M., Mohamed, F.A. and Ibrahim, M.S. 2017. Pathogenicity and characteriztion of Streptococcosis in Egyptian Nile tilapia (Oreochromis niloticus) in Kafr Elshikh Governorate. AJVS 52(1), 173–180. Sarkar, A., Mousumi, S. and Roy, P. 2012. Identification and typing of Aeromonas hydrophila through 16S rDNA-PCR fingerprinting. Aquacul. Res. Develop. 3(4), 142–145. Sarkar, A., Saha, M. and Roy, P. 2013. Detection of 232bp virulent gene of pathogenic Aeromonas hydrophila through PCR based technique: (A Rapid Molecular Diagnostic Approach). Adv. Microbiol. 3, 83–87. Schaperclaus, W., Kulow, H. and Schreckenbach, K. 1992. Infections abdominal dropsy. In Fish disease. Ed., Schaperclaus, W. Berlin, Germany: Akademie-Verlag, vol. 1, pp: 401–458. Sekar, V., Santiago, T., Vijayan, K., Alavandi, S., Raj, V., Rajan, J., Sanjuktha, M. and Kalaimani. N. 2008. Involvement of Enterobacter cloacae in the mortality of the fish, Mugil cephalus. Lett. Appl. Microbiol. 46(6), 667–672. Shaheen, A., Seisay, M. and Nouala, S. 2013. An industry assessment of tilapia farming in Egypt. African Union, International Bureau for Animal Resources (AU-IBAR). Small, B. and Bilodeau, A. 2005. Effects of cortisol and stress on channel catfish (Ictalurus punctatus) pathogen susceptibility and lysozyme activity following exposure to Edwardsiella ictaluri. Gen. Comp. Endocrinol. 142(1-2), 256–262. Soliman, M.K., Ellakany, H.F. , Gaafar, A.Y., Elbialy, A.K., Zaki, M.S. and Younes, A.M. 2014. Epidemiology and antimicrobial activity of methicillin-resistant Staphylococcus aureus (MRSA) isolated from Nile tilapia (Oreochromis niloticus) during an outbreak in Egypt. Life Sci. J. 11(10), 1245–1252. Soliman, N.F. and Yacout, D.M.M. 2016. Aquaculture in Egypt: status, constraints and potentials. Aquac. Int. 24(5), 1201–1227. Stratev, D. and Odeyemi, O.A. 2016. Antimicrobial resistance of Aeromonas hydrophila isolated from different food sources: a mini-review. J. Inf. Public. Health. 9(5), 535–544. Tarr, C.L., Patel, J.S., Puhr, N.D., Sowers, E.G., Bopp, C.A. and Strockbine, N.A. 2007. Identification of Vibrio isolates by a multiplex PCR assay and rpoB sequence determination. J. Clin. Microbiol. 45(1), 134–140. Thompson, R.C., Olsen, Y., Mitchell, R.P., Davis, A., Rowland, S.J. and John, A.W.G. 2004. Lost at sea: where is all the plastic? Science 304(5672), 838. Tsai, M.A., Ho, P.Y., Wang, P.C., E, Y.J., Liaw, L.L. and Chen, S.C. 2012. Development of a multiplex polymerase chain reaction to detect five common Gram-negative bacteria of aquatic animals. J. Fish. Dis. 35, 489–495. Umesha, D., Srinivasa-Rao, P., Pani-Prasad, K., Reddy, A.K., SrinivasK, N. 2011. Aerolysin and haemolysin virulence genes of Aeromonas hydrophila isolated from diseased ornamental freshwater Oscar fish and goldfish by polymerase chain reaction. Int. J. Adv. Sci. Technol. 3(1), 82–89. Vazirzadeh, A., Jalali, S. and Farhadi, A. 2019. Antibacterial activity of Oliveria decumbens against Streptococcus iniae in Nile tilapia (Oreochromis niloticus) and its effects on serum and mucosal immunity and antioxidant status. Fish. Shellfish. Immunol. 94, 407–416. Walakira, J., Akoll, P., Engole, M., Sserwadda, M., Nkambo, M., Namulawa, V., Kityo, G., Musimbi, F., Abaho, I. and Kasigwa, H. 2014. Common fish diseases and parasites affecting wild and farmed tilapia and catfish in Central and Western Uganda. Uganda. J. Agric. Sci. 15(2), 113–125. Wedekind, C., Gessner, M.O., Vazquez, F., Maerki, M. and Steiner, D. 2010. Elevated resource availability sufficient to turn opportunistic into virulent fish pathogens. Ecology 91(5), 1251–1256. Yambot, A. (1998). Isolation of Aeromonas hydrophila from Oreochromis niloticus during fish disease outbreaks in the Philippines. Asian. Fish. Sci.10, 347–354. Yardimci, B. and Aydin, Y. 2011. Pathological findings of experimental Aeromonas hydrophila infection in Nile tilapia (Oreochromis niloticus). Ankara. Üniv. Vet. Fak. Derg. 58, 47–54. Ye, X., Li, J., Lu, M., Deng, G., Jiang, X., Tian, Y., Quan, Y. and Jian, Q. 2011. Identification and molecular typing of Streptococcus agalactiae isolated from pond-cultured tilapia in China. Fish. Sci. 77(4), 623–632. Younes, A.M., Gaafar, A.Y., Abu-Bryka, A.Z., Mohamed, L.A. and Bayoumy E.M. 2019. Genotyping and pathogenicity of Streptococcus iniae strains recovered from cultured Oreochromis niloticus at Kafr El-Shiekh Governorate, Egypt. Egypt. J. Aquat. Biol. Fish. 23(3), 467–474. Youssuf, H., Abdel Gawad, E.A., El Asely, A.M., Elabd, H., Matter, A.F., Shaheen, A.A. and Abbass, A.A. 2020. Insight into summer mortality syndrome in farmed Nile tilapia (Oreochromis niloticus) associated with bacterial infection. Benha. Vet. Med. J. 39(1), 111–118. Zheng, D., Mai, K., Liu, S., Limin, C., Liufu, Z., Xu, W., Tan, B. and Zhang, W. 2004. Effect of temperature and salinity on virulence of Edwardsiella tarda to Japanese flounder, Paralichthys olivaceus (Temminck et Schlegel). Aqua. Res. 35, 494–500. Zlotkin, A., Hershko, H. and Eldar, A. 1998. Possible transmission of Streptococcus iniae from wild fish to cultured marine fish. Appl. Environ. Microbiol. 64, 4065–4067. | ||
| How to Cite this Article |
| Pubmed Style Magouz FI, Moustafa EM, Abo-remela EM, Halawa MR, Barakaat PM, Omar AA. Summer mortality syndrome bacterial pathogens in farmed Nile tilapia (Oreochromis niloticus). Open Vet. J.. 2024; 14((1) (Zagazig Veterinary Conference)): 53-69 . doi:10.5455/OVJ.2024.v14.i1.7 Web Style Magouz FI, Moustafa EM, Abo-remela EM, Halawa MR, Barakaat PM, Omar AA. Summer mortality syndrome bacterial pathogens in farmed Nile tilapia (Oreochromis niloticus). https://www.openveterinaryjournal.com/?mno=174702 [Access: January 11, 2026]. doi:10.5455/OVJ.2024.v14.i1.7 AMA (American Medical Association) Style Magouz FI, Moustafa EM, Abo-remela EM, Halawa MR, Barakaat PM, Omar AA. Summer mortality syndrome bacterial pathogens in farmed Nile tilapia (Oreochromis niloticus). Open Vet. J.. 2024; 14((1) (Zagazig Veterinary Conference)): 53-69 . doi:10.5455/OVJ.2024.v14.i1.7 Vancouver/ICMJE Style Magouz FI, Moustafa EM, Abo-remela EM, Halawa MR, Barakaat PM, Omar AA. Summer mortality syndrome bacterial pathogens in farmed Nile tilapia (Oreochromis niloticus). Open Vet. J.. (2024), [cited January 11, 2026]; 14((1) (Zagazig Veterinary Conference)): 53-69 . doi:10.5455/OVJ.2024.v14.i1.7 Harvard Style Magouz, F. I., Moustafa, . E. M., Abo-remela, . E. M., Halawa, . M. R., Barakaat, . P. M. & Omar, . A. A. (2024) Summer mortality syndrome bacterial pathogens in farmed Nile tilapia (Oreochromis niloticus). Open Vet. J., 14 ((1) (Zagazig Veterinary Conference)), 53-69 . doi:10.5455/OVJ.2024.v14.i1.7 Turabian Style Magouz, Fawzy I., Eman M. Moustafa, Etab M. Abo-remela, Marwa R. Halawa, Passant M. Barakaat, and Amira A. Omar. 2024. Summer mortality syndrome bacterial pathogens in farmed Nile tilapia (Oreochromis niloticus). Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 53-69 . doi:10.5455/OVJ.2024.v14.i1.7 Chicago Style Magouz, Fawzy I., Eman M. Moustafa, Etab M. Abo-remela, Marwa R. Halawa, Passant M. Barakaat, and Amira A. Omar. "Summer mortality syndrome bacterial pathogens in farmed Nile tilapia (Oreochromis niloticus)." Open Veterinary Journal 14 (2024), 53-69 . doi:10.5455/OVJ.2024.v14.i1.7 MLA (The Modern Language Association) Style Magouz, Fawzy I., Eman M. Moustafa, Etab M. Abo-remela, Marwa R. Halawa, Passant M. Barakaat, and Amira A. Omar. "Summer mortality syndrome bacterial pathogens in farmed Nile tilapia (Oreochromis niloticus)." Open Veterinary Journal 14.(1) (Zagazig Veterinary Conference) (2024), 53-69 . Print. doi:10.5455/OVJ.2024.v14.i1.7 APA (American Psychological Association) Style Magouz, F. I., Moustafa, . E. M., Abo-remela, . E. M., Halawa, . M. R., Barakaat, . P. M. & Omar, . A. A. (2024) Summer mortality syndrome bacterial pathogens in farmed Nile tilapia (Oreochromis niloticus). Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 53-69 . doi:10.5455/OVJ.2024.v14.i1.7 |