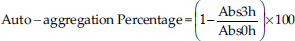| Research Article | ||
Open Vet. J.. 2024; 14(2): 716-729 Open Veterinary Journal, (2024), Vol. 14(2): 716–729 Original Research Genomic characterization and probiotic potential of lactic acid bacteria isolated from feces of guinea pig (Cavia porcellus)José Goicochea-Vargas1,2*, Max Salvatierra-Alor2, Fidel Acosta-Pachorro1, Wilson Rondón-Jorge1, Arnold Herrera-Briceño3, Edson Morales-Parra4 and Eric Mialhe51Facultad de Medicina Veterinaria y Zootecnia, Universidad Nacional Hermilio Valdizan, Huánuco, Peru 2Laboratorio de Biotecnología Molecular, Unidad Central de Laboratorios, Universidad Nacional Hermilio Valdizan, Huánuco, Peru 3Centros de Producción Canchán y Kotosh, Universidad Nacional Hermilio Valdizan, Huánuco, Peru 4Centro de Información y Educación para la Prevención del Abuso de Drogas—CEDRO, Lima, Peru 5INCABIOTEC SAC, Tumbes, Peru *Corresponding Author: José F. Goicochea Vargas. Facultad de Medicina Veterinaria y Zootecnia, Universidad Nacional Hermilio Valdizan, Huánuco, Peru. Email: jgoicochea [at] unheval.edu.pe Submitted: 10/12/2023 Accepted: 23/01/2023 Published: 29/02/2024 © 2024 Open Veterinary Journal
AbstractBackground: Presently, there exists a growing interest in mitigating the utilization of antibiotics in response to the challenges emanating from their usage in livestock. A viable alternative strategy encompasses the introduction of live microorganisms recognized as probiotics, exerting advantageous impacts on the immune system and nutritional aspects of the host animals. Native lactic acid bacteria, inherently possessing specific properties and adaptive capabilities tailored to each animal, are deemed optimal contenders for probiotic advancement. Aim: In the current investigation, microorganisms exhibiting probiotic potential were isolated, characterized, and identified from the fecal samples of guinea pigs (Cavia porcellus) belonging to the Peruvian breed. Methods: The lactic acid bacteria isolated on Man, Rogosa, and Sharpe agar underwent Gram staining, catalase testing, proteolytic, amylolytic, and cellulolytic activity assays, low pH tolerance assessment, hemolytic evaluation, antagonism against Salmonella sp., determination of autoaggregation and coaggregation capacity, and genotypic characterization through sequencing of the 16S rRNA gene. Results: A total of 33 lactic acid bacteria were isolated from the feces of 30 guinea pigs, also 10 isolates were selected based on Gram staining and catalase testing. All strains exhibited proteolytic activity, while only one demonstrated amylolytic capability, and none displayed cellulase activity. These bacteria showed higher tolerance to pH 5.0 and, to a lesser extent, to pH 4.0. Furthermore, they exhibited antagonistic activity against Salmonella sp. Only two bacteria demonstrated hemolytic activity, and were subsequently excluded from further evaluations. Subsequent assessments revealed autoaggregation capacities ranging from 4.55% to 23.19%, with a lesser degree of coaggregation with Salmonella sp. ranging from 3.53% to 8.94% for the remaining eight bacterial isolates. Based on these comprehensive tests, five bacteria with notable probiotic potential were identified by molecular assays as Leuconostoc citreum, Enterococcus gallinarum, Exiguobacterium sp., and Lactococcus lactis. Conclusion: The identified bacteria stand out as promising probiotic candidates, deserving further assessment in Peruvian breed guinea pigs. This exploration aims to enhance production outcomes while mitigating the adverse effects induced by pathogenic microorganisms. Keywords: Lactic acid bacteria, Enzymatic capacity, Salmonella sp., PCR, 16S rRNA. IntroductionGuinea pigs, originally domesticated in Peru and subsequently introduced to various regions spanning South America, the Caribbean, Europe, and the United States (Lord et al., 2020), hold significance in breeding endeavors owing to the exceptional quality of their meat. This meat is characterized by its elevated protein content and low fat composition, rendering it an appealing choice for human consumption (Enríquez, 2019). Integral to their productive enterprise is the meticulous management of nutrition, primarily centered around green forage. Nevertheless, alternative nutritional sources, including balanced feed, vitamins, and high-protein flours, have been integrated into their dietary regimen (Reynaga et al., 2020; Herrera et al., 2022). In tandem with nutritional considerations, scrutiny has been directed toward the intricate microbial communities inhabiting the gastrointestinal tract of diverse animal species, given their profound influence on host physiological processes (Esser et al., 2019; Frias et al., 2023). Within the intestinal milieu of guinea pigs, the discerned microbial diversity is principally constituted by phyla encompassing Bacteroidetes, Firmicutes, Fibrobacteres, Proteobacteria, Actinobacteria, Verrucomicrobia, and Tenericutes (Crowley et al., 2017; Palakawong Na Ayudthaya et al., 2019). The nuanced distribution of these phyla is contingent upon the specific intestinal segment under examination (Tang et al., 2022). Within this microbial consortium, select microorganisms demonstrate the capability to extracellularly produce enzymes and metabolites, thereby exerting influence over nutrient absorption dynamics within the host’s intestinal milieu (Solden et al., 2018; Oliphant and Allen-Vercoe, 2019). Furthermore, these microorganisms intricately engage with the host’s immune system, modulating defensive responses against a spectrum of pathogens (Zheng et al., 2020). The confluence of technological advancements and intensified guinea pig production has brought to light profound sanitary challenges precipitated by microbial pathogens such as Salmonella sp. and Escherichia coli. These pathogens give rise to enterohepatic lesions, while Streptococcus sp. is implicated in respiratory afflictions, and to a lesser extent, Klebsiella sp. and Bordetella sp. (Morales, 2017; Angulo-Tisoc et al., 2021). These microbial agents contribute significantly to elevated mortality rates and suboptimal growth in guinea pigs (Obregón et al., 2018; Bazán et al., 2019; Angulo-Tisoc et al., 2021). Despite the widespread practice of administering antibiotics by producers to mitigate the deleterious effects of these infections, empirical evidence underscores the persistent presence of these antibiotics in guinea pig meat, as well as in the liver and kidneys (Ampuero-Riega and Morales-Cauti, 2021). Furthermore, there is a discernible surge in pathogen resistance to antibiotics, as substantiated by research findings (Quesada et al., 2016). A viable alternative to antibiotic administration is the integration of probiotics into animal diets. Probiotics, living microorganisms recognized for their positive health impact in both human and animal contexts (Das et al., 2022), operate through mechanisms such as intestinal colonization, normalization of microbial communities, competitive exclusion of pathogenic counterparts, production of short-chain branched fatty acids, intestinal mucus synthesis, and modulation of the immune system, among other factors (Plaza-Diaz et al., 2019). The application of probiotics has yielded substantial enhancements in the production performance of various animals, marked by elevated growth rates, improved feed conversion efficiency, and diminished mortality due to infectious diseases (Bhogoju and Nahashon, 2022). In the specific context of guinea pig production, probiotic supplementation has been associated with enhanced feed conversion rates and immune stimulation levels akin to those achieved using growth-promoting antibiotics in animals afflicted with Salmonella sp. (Saldarriaga, 2018; Carcelén et al., 2021).Preeminent among microorganisms employed as probiotics are those belonging to the group of lactic acid bacteria. This category encompasses gram-positive facultative anaerobic cocci and bacilli, distinguished by their lack of catalase activity, non-sporulation, and proficiency in lactic acid production (Pachla et al., 2018; Bintsis, 2018). Based on their metabolic pathways, they are stratified into homofermentative types, which ferment sugars to yield lactic acid, and heterofermentative types, generating lower acid quantities but producing carbon dioxide, ethanol, and mannitol (Moon et al., 2018; Abedi and Hashemi, 2020). These microorganisms are deemed promising probiotics due to their capabilities in organic acid and bacteriocin production, impeding the proliferation of pathogens. In addition, they exhibit the synthesis of host-beneficial vitamins, secretion of immune-enhancing exopolysaccharides, and production of antioxidant substances (Wang et al., 2021). Noteworthy genera renowned for their probiotic potential include Enterococcus, Pediococcus, Lactobacillus, Streptococcus, and Leuconostoc (Fijan, 2014). In light of the aforementioned, there is a notable emphasis on the imperative to continue research on novel lactic acid bacteria possessing probiotic potential, particularly for their application in guinea pig rearing with the aim of improving production outcomes. Therefore, the primary objective of this study was to isolate and characterize native strains of lactic acid bacteria derived from guinea pig feces. The study further sought to evaluate the in vitro probiotic potential of these strains and molecularly identify the selected candidates. Materials and MethodsCollection of guinea pig fecal samplesGuinea pig fecal samples were procured in February 2023 at the Kotosh Production Center of the Hermilio Valdizan National University (UNHEVAL), situated in Kotosh City, Huánuco Province, within the Huánuco Region, Peru. A total of 40 guinea pigs of the Peruvian breed, weighing between 300 and 400 grams, were selected for the study. These animals were evenly distributed among four ponds, each containing 10 individuals. A 2-week observation period preceded the sampling process to ensure the absence of disease signs, and no antibiotics were administered during this period. Pond two, displaying signs of illness, was consequently excluded from the study. During the observation period, the guinea pigs were acclimated to a diet consisting of green forage, comprising corn husks, alfalfa, and hydroponically grown barley, with no water supplementation. The animals were exposed to a lighting regimen of 12 hours of light and 12 hours of darkness, maintained under ambient temperature conditions. Approximately 10 grams of fresh feces were collected from each pond using sterile forceps and promptly placed into previously sterilized tubes. The tubes containing fecal samples were then placed in a cooler box with refrigerant gel to maintain a temperature range between 2°C and 8°C during transportation to the Molecular Biotechnology Laboratory at UNHEVAL. This controlled temperature ensured the preservation of sample integrity for subsequent processing (Serrano et al., 2020). Isolation of lactic acid bacteriaThe collected fecal samples were cleaned and processed within a Class II biosafety cabinet (Biobase, China) using surgical blades to extract the superficial portion of the feces, subsequently placing them in Petri dishes for each of the three designated groups. One gram of fecal surface was taken and diluted in 50 ml of sterile saline solution (NaCl 0.9%). Subsequently, 1 ml of the prepared solution underwent serial dilutions to a value of 10-4. One hundred microliters of each dilution were spread on Man, rogosa, and sharpe (MRS) agar plates (Hi-Media, India), a medium specific for lactic acid bacteria. These plates were then incubated under microaerophilic conditions at 37°C for 24 hours. Bacterial colonies obtained were further purified through sub-culturing on MRS agar plates, followed by an additional incubation at 37°C for 20 hours under microaerophilic conditions to ensure the attainment of pure colonies. Finally, 10–12 colonies were selected from each group (designated as G1, G3, and G4) for subsequent evaluations. Biochemical selection testsThe pure bacterial isolates underwent characterization through Gram staining and a catalase activity test. For the Gram staining, a Gram staining kit (LabiFarma, Peru) was employed. The process involved initially staining the bacterial smear, fixed by prior flame fixation, with crystal violet for 1 minutes. Subsequently, Lugol’s solution was added for 1 minutes, followed by acetone alcohol for 15 seconds, and safranin for 1 minutes. Distilled water rinses were performed between each treatment. The slides were observed under a microscope at 1,000× magnification using immersion oil (Jain et al., 2020). Catalase activity was assessed by adding three drops of 15% hydrogen peroxide (JTBaker, USA) onto a bacterial smear on a glass slide. The appearance of bubbles was considered indicative of catalase enzyme activity (Castillo et al., 2022). For subsequent tests, Gram-positive bacteria with negative catalase activity were selected. Evaluation of enzymatic activityProteolytic activityThe proteolytic activity of the isolates was assessed using Tryptic Soy Agar (TSA) agar plates (Difco, USA) supplemented with 1.5% skim milk (Difco, USA). Before testing, a culture of each isolate was prepared in MRS broth (Hi-Media, India) and incubated at 37°C for 20 hours under microaerophilic conditions. Subsequently, the bacterial concentration was measured using a Nanodrop Onec UV-visible spectrophotometer (Thermo Scientific, USA) set at 650 nm and adjusted to a final bacterial density of 1.0 optical density (OD 650). Next, 5 μl of each bacterial culture was inoculated onto TSA plates supplemented with skim milk, in triplicate. The plates were allowed to dry and then incubated at 37°C for 48 hours under microaerophilic conditions. The formation of a clear halo around the bacterial colony indicated proteolytic activity. Following the methodology outlined by Fitriyanto et al. (2020), the diameter of the halo and the bacterial colony were measured to determine the proteolytic index using the following formula:
Amylolytic activityThe assessment of amylolytic activity was conducted on nutrient agar plates (Hi-Media, India) supplemented with 1% soluble starch (Millipore, Germany). From bacterial cultures in MRS broth, 5 μl of each culture adjusted to a concentration of 1 OD 650 were inoculated in triplicate onto plates with nutrient agar and soluble starch. The plates were allowed to dry and then incubated at 37°C for 48 hours under microaerophilic conditions. Subsequently, 3 ml of a Lugol’s solution (Sigma-Aldrich, Germany) was added to each plate and allowed to stand for 5–10 minutes at room temperature. The appearance of a halo around the bacterial colony was considered an indicator of starch hydrolysis. Furthermore, the diameter of the halo and bacterial colony was measured to determine the amylolytic index using the following formula:
Cellulolytic activityThe cellulolytic activity of the isolates was assessed on nutrient agar plates supplemented with 1% Carboxymethyl cellulose (Spectrum Chemical, China). For each bacterial culture, 5 μl adjusted to a concentration of 1.0 OD650 were inoculated in triplicate onto plates with nutrient agar and carboxymethyl cellulose. The plates were allowed to dry and then incubated at 37°C for 48 hours under microaerophilic conditions. To reveal cellulose degradation, a 1.0% Congo red solution (Merck, Germany) was added, and the plates were allowed to stand for 15 minutes. The excess solution was removed, and a 0.1 M NaCl solution was added, left to stand for another 15 minutes, and the excess was removed. Finally, the plate was covered with 2% acetic acid for 10 seconds (Alcívar-Vega and Vera-Vargas, 2013; Anguiano-Aguilar, 2019). The presence of a clear halo around the colonies indicates cellulolytic activity. Evaluation of probiotic capacityTolerance to low pHSterile MRS broth was prepared in tubes with 4.5 ml, and the pH was adjusted to 6.5, 5.0, and 4.0 using concentrated HCl. Simultaneously, an inoculum of each isolate were cultured in MRS broth at 37°C for 20 hours under microaerophilic conditions. Subsequently, 0.5 ml (10% v/v) of each culture was added to the tubes with MRS broth at the adjusted pH levels, with triplicate inoculations. The tubes were then incubated for 18 hours at 37°C under microaerophilic conditions. Following incubation, bacterial concentration was measured by OD at 650 nm using a UV-visible spectrophotometer. Comparisons were made between bacterial concentrations at pH 5.0 and 4.0, adjusted with the concentration obtained at pH 6.5 for each bacterium. Antagonistic activity against Salmonella sp.To assess antagonistic activity, we employed a modified agar overlay method inspired by Halder et al. (2017). Inocula of each bacterial isolate was cultured in MRS broth and incubated at 37°C for 20 hours under microaerophilic conditions. The OD for each microbial isolate was adjusted to 1 OD650 using a spectrophotometer. Subsequently, 5 μl of each isolate was inoculated in triplicate onto MRS agar plates. These plates were covered with Mueller Hinton agar (Condalab, Spain) containing 0.8% agar, pre-mixed with 106 CFU/ml of Salmonella sp. After drying, the plates were incubated at 37°C for 48 hours under microaerophilic conditions to observe the presence or absence of an inhibition halo formed around the bacterial colony. Hemolytic activityThe hemolytic activity of bacterial isolates was assessed by triplicate inoculation of 5 μl of bacterial culture adjusted to a concentration of 1.0 OD650 onto azide blood agar plates (Microgen, India) containing 5% human blood. The plates were incubated under microaerophilic conditions at 37°C for 72 hours. The presence of hemolytic activity was associated with the formation of a clear halo around bacterial colonies (Kousha et al., 2022). Auto-aggregation assayThe selected bacteria were inoculated into MRS broth and incubated at 37°C under microaerophilic conditions for 20 hours. After centrifugation at 3,000 g for 10 minutes, the supernatant was discarded. The bacterial pellet was washed twice with a PBS 1× buffer (pH 7.2) and resuspended in the same solution. The bacterial concentration was adjusted to an OD650 of 0.25 ± 0.05. Subsequently, 3 ml of bacterial suspensions were aliquoted in triplicate and incubated at 37°C for 3 hours. The absorbance was measured at 650 nm. The percentage of auto-aggregation was expressed as follows:
where Abs3 hours represents the absorbance at 650 nm after 3 hours of incubation, and Abs0h represents the absorbance at 650 nm at the start of incubation (0 hour) (Tuo et al., 2013). Co-aggregation assayThe selected bacteria were cultured in MRS broth and incubated at 37°C under microaerophilic conditions for 20 hours. In addition, an aliquot of Salmonella sp. was cultured in TSB broth (Difco, USA) and incubated at 37°C for 20 hours. Suspensions of lactic acid bacteria and Salmonella sp. were prepared in PBS 1× as described in the auto-aggregation assay, considering an OD650 of 0.29 ± 0.05 for Salmonella sp. Subsequently, 1 ml of the suspension of each evaluated bacterium was mixed with 1 ml of Salmonella sp. suspended in PBS 1×. The mixture underwent vortex agitation and was incubated at 37°C for 3 hours. The absorbance was then measured at 650 nm. The percentage of co-aggregation was calculated as follows:
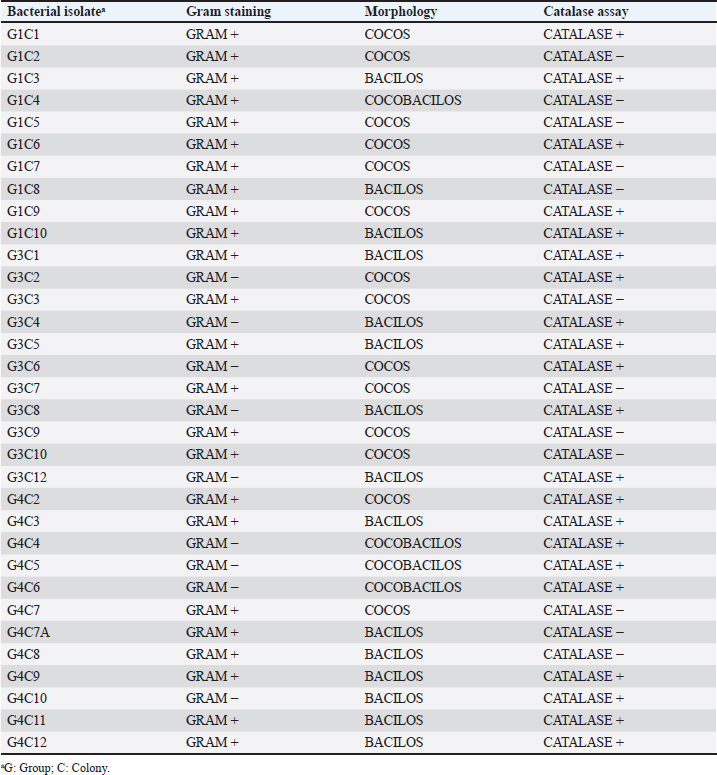
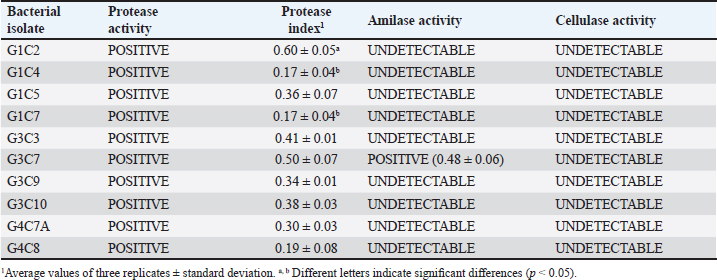
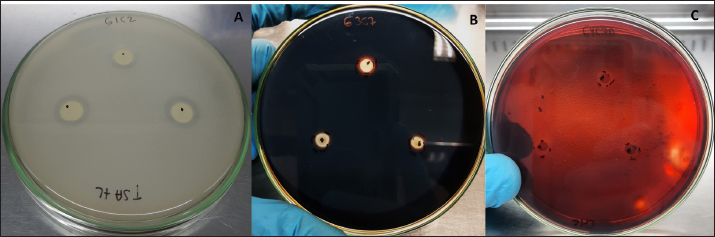
where AbsSalm and Absacid correspond to the absorbance at 650 nm of the Salmonella sp. and the lactic acid bacteria suspension at the beginning of the assay. Absmixture corresponds to the absorbance at 650 nm of the mixture of Salmonella sp. with lactic acid bacteria after 3 hours of the assay (Tuo et al., 2013). Bacterial molecular identification using 16S rRNA markerThe selected bacterial isolates from the previous tests were identified using molecular assays targeting a fragment of the 16S rRNA gene. Bacterial cultures were grown in MRS broth for 20 hours at 37°C under microaerophilic conditions, and their concentration was adjusted to 1.0 OD650. Bacteria were then centrifuged at 5,000 g for 10 minutes, and the resulting pellet was resuspended in saline solution (NaCl 0.9%). Genomic DNA (gDNA) was extracted using the GeneJet Genomic DNA Purification Kit (Thermo Scientific, USA), following the manufacturer’s recommended procedures (Ahmed et al., 2021). The quantity and purity of the obtained gDNA were verified using the spectrophotometer NanoDrop OneC (Thermo Scientific, USA). The bacterial genomic DNA’s 16S rRNA gene fragment was amplified using universal primers 27F: 5′ AGAGTTTGATCCTGGCTCAG 3′ and 1492R: 5′ GGTTACCTTGTTACGACTT 3′ at a concentration of 0.24 uM. The amplification was carried out through Polymerase Chain Reaction (PCR), employing 1U of Maximo Taq DNA polymerase (Geneon, Germany) and 0.2 mM dNTPs (Geneaid) in a 50 ul reaction volume. PCR was performed using a SimpliAmp thermocycler (Applied Biosystem, USA), initiating with an initial denaturation step at 95°C for 5 minutes. This was followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 54°C for 40 seconds, and extension at 72°C for 1 minutes. An additional extension step at 72°C for 7 minutes concluded the cycling, followed by a final step at 4°C. The presence of PCR products was confirmed by electrophoresis on 1.5% agarose gels (Geneon, Germany) alongside a 100 bp DNA ladder (Geneaid Biotech, Taiwan). The PCR products were analyzed by 1.5 % agarose gel electrophoresis using a 1× buffer Tris-acetate-EDTA also used for the immersion of gel. Electrophoresis was run at 90V for 40 minutes in a Sub Cell GT horizontal electrophoresis chamber (BioRad, USA) connected to a PowerPac HC power supply (BioRad, USA). Gel visualization was performed using a GelDoc Go Imaging System (Biorad, USA). PCR products with a band size of approximately 1,500 bp were purified and sent for sequencing to Macrogen Inc., (Chile) using the ABI 3730XL DNA sequencer (Applied Biosystem, USA). Sequencing was carried out with primers targeting the V3-V4 region: 16S_V3-V4_IlluminaF (CCTACGGGGNGGCWGCAG) and 16S_V3-V4_IlluminaR (GACTACHVGGGTATCTAATCC). The generated sequences underwent analysis using the software program DNA Baser Sequence Assembler v5.21.0 (Heracle BioSoft, Romania) to confirm base assignments and sequence assembly. In addition, the Blastn free program (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_SPEC=GeoBlast&PAGE_TYPE=BlastSearch) was employed for the identification of genera and/or species corresponding to the obtained sequences. Phylogenetic relationships were inferred using the program MEGA 11 (Tamura et al., 2021). Phylogenetic trees were constructed using the maximum likelihood method with the Kimura 2-parameter model, incorporating a gamma distribution of substitution rates among sites and 1,000 bootstrap replicates. Sequences obtained in this study were used alongside 14 16S rRNA gene sequences retrieved from the NCBI GenBank database. Statistical analysisSoftware Microsoft Excel v.2312 (Microsoft Corporation, USA) was used to create tables and comparative graphs. Statistical tests were conducted using the program IBM SPSS Statistics v.26 (IBM Corp, USA). The non-parametric Kruskal-Wallis test (proteolytic activity, pH tolerance, autoaggregation, and coaggregation) for independent samples was employed with a 95% confidence interval. Multiple comparisons were conducted using the Dunn test with a Bonferroni correction for the level of significance. Ethical approvalNot needed because the animal manipulation was only for the collection of feces, the principles of animal welfare according to the World Organization for Animal Health (founded as OIE) were complied with and the animals were not sacrificed in this study. ResultsAnimals were randomly distributed in groups of 10 animals per pond to reduce stress due to high densities. The feces collected from each pond represented 03 replicates for the isolation bacteria that maximize the probabilities of obtaining acid lactic microorganisms with different morphologies for the following evaluations. A total of 33 pure bacterial isolates were obtained, distributed among Group G1 (10 isolates), Group G3 (11 isolates), and Group G4 (12 isolates), as detailed in Table 1. Gram staining characterization revealed that 73% exhibited a Gram-positive pattern, with 27% showing Gram-negative characteristics. Microscopic examination of all isolates identified cocci (42%), rods or bacilli (45%), and coccobacilli (13%). Notably, within the Gram-positive bacterial group, similar percentages were observed for both cocci and bacilli. Furthermore, catalase testing indicated that 64% of the isolates were catalase-positive, while 36% exhibited catalase-negative activity. From the initial characterization, ten acid lactic bacterial isolates were chosen for further evaluation, which were Gram-positive organisms, catalase negative, mesophilic, and rod or spherical shaped. The selected ones are: G1C2, G1C4, G1C5, G1C7, G3C3, G3C7, G3C9, G3C10, G4C7A, and G4C8. Isolates G1C8 and G4C7 were not considered due to their low or negligible growth in the conducted cultures. The potential for extracellular enzymatic activity of each selected microbial isolate was investigated using selective agar for protease, amylase, and cellulase activities. In all three cases, the formation of a clear halo in the agar around bacterial colonies indicated enzymatic activity. The results are presented in Table 2. The assay for proteolytic activity revealed that all bacterial isolates exhibited extracellular protease activity, forming a halo of protein degradation in the agar around the colony. The calculated proteolytic indices ranged from 0.17 to 0.60, indicating significant differences among them (p=0.001). Isolates G1C2 and G3C7 demonstrated higher proteolytic activity, while G1C4 and G1C7 showed lower activity. However, significant differences were observed only between G1C2 and G1C4 (p=0.024) and G1C7 (p=0.034). In this study, amylase activity was exclusively identified in the G3C7 isolate, demonstrating an amylolytic index of 0.48 ± 0.06 (Table 2). Conversely, cellulase activity remained undetected in the assessed bacterial isolates (Fig. 1C). Table 1. Gram staining, bacterial morphology, and catalase activity tests for isolates from guinea pig feces.
Table 2. Assessment of protease, amylase, and cellulase enzymatic activity in selected bacterial isolates.
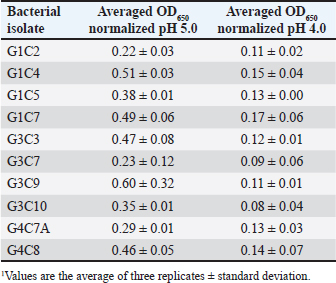
Fig. 1. Extracellular enzymatic activity of selected isolates. A) Isolate G1C2 exhibiting the highest protease activity, B) Isolate G3C7 displaying amylase activity, and C) Isolate G4C7A with negative cellulase activity. Table 3. Growth measured in OD650 of microbial isolates in MRS broth at pH 5.0 and 4.01 normalized with respect to growth at pH 6.5.
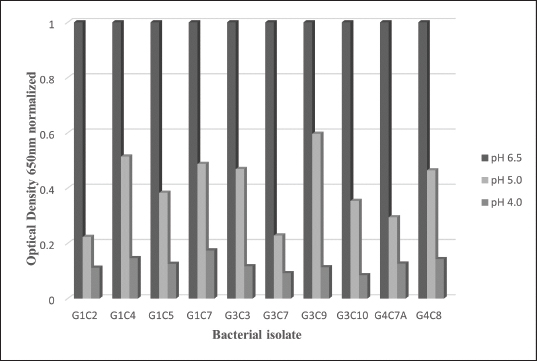
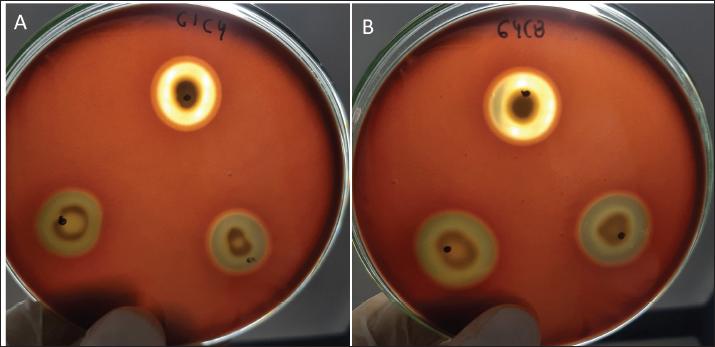
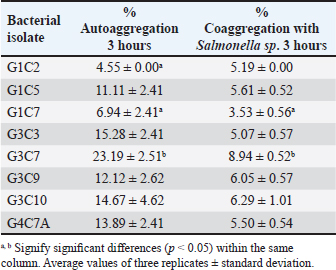
Probiotic capacity assaysTolerance to low pH levelsOne of the defining features of microorganisms with probiotic potential is their ability to resist growth in environments with low pH levels, facilitating their survival in the digestive tract of animals (Bernatek et al., 2022; Mendonça et al., 2022). The OD650 growth data at pH 6.5, 5.0, and 4.0 were normalized to the growth at pH 6.5 for comparative analyses (Table 3). The results revealed a significant reduction in bacterial growth due to the decrease in pH, particularly evident in cultures at pH 4.0 (p < 0.05) compared to cultures at pH 6.5 for each strain evaluated. Differences in growth at pH 5.0 compared to pH 6.5 were also observed (p > 0.05). Isolates demonstrating higher tolerance at pH 5.0 included G3C9 (0.6 ± 0.32), followed by G1C4 (0.51 ± 0.03) and G1C7 (0.49 ± 0.06), while G1C2 exhibited lower tolerance (0.22 ± 0.03) (p > 0.05). Consistently, G1C4 and G1C7 displayed greater tolerance at pH 4.0 compared to other isolates (p > 0.05). Antagonistic activity against Salmonella sp.All the assessed microorganisms exhibited inhibition zones against the pathogenic bacterium Salmonella sp., as demonstrated by the clear halos observed in the superficial agar layer. Hemolytic capacityAs for this investigation, isolates G1C4 and G4C8 (Fig. 3) displayed distinct halos with a slightly greenish hue on the blood agar, leading to their exclusion from subsequent assays. For the remaining bacteria, no halos were discerned in the agar medium. Autoaggregation and coaggregation capacity against Salmonella sp.Eight isolates were assessed to determine their autoaggregation capacity at 3 hours (Table 4). The highest autoaggregation percentage was observed in isolate G3C7 at 23.19% ± 2.51%, compared to 4.55% ± 0.00% in G1C2 and 6.94% ± 2.41% in G1C7, revealing significant differences (p < 0.05). Similarly, the eight bacterial isolates were examined to ascertain their coaggregation capacity with the pathogen Salmonella sp. The highest coaggregation capacity was found to be 8.94% ± 0.52% in G3C7, which was significantly different from 3.53% ± 0.56% in G1C7 (p < 0.05).
Fig. 2. Bar chart illustrating the tolerance of bacterial isolates to pH 5.0 and 4.0 expressed in OD values at 650 nm normalized with respect to growth at pH 6.5. (*) indicates significant differences.
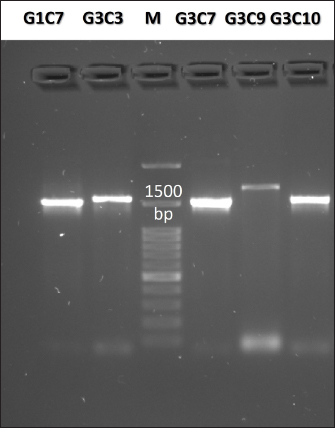
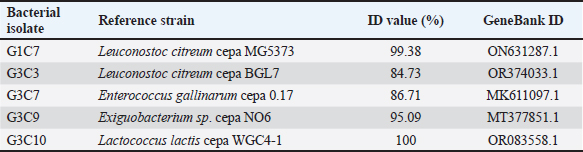
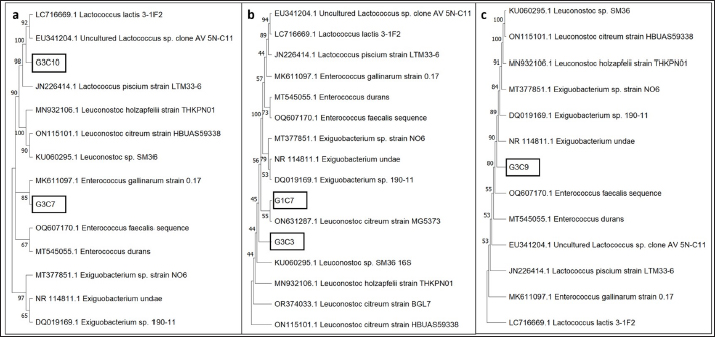
Fig. 3. Illustration of the hemolytic capacity of bacterial isolates, highlighting a greenish halo in the cases of (A) G1C4 and (B) G4C8. Molecular identification using the 16S rRNA geneConsidering the probiotic capacity criteria evaluated earlier, isolates G1C7, G3C3, G3C7, G3C9, and G3C10 were selected for molecular identification. Positive amplicons of approximately 1,500 bp were obtained for all 05 isolates after agarose gel migration (Fig. 4). The obtained 16S rRNA gene sequences for isolates G1C7 and G3C3 correspond to Leuconostoc citreum, with identity percentages of 99.38% and 84.73%, respectively. Isolates G3C7, G3C9, and G3C10 were identified as Enterococcus sp., Exiguobacterium sp., and Lactococcus lactis with identity percentages of 86.71%, 99.38%, and 100%, respectively (Table 5), and an E-value very close to zero. In addition, phylogenetic trees were constructed for isolates G3C7 and G3C10, one for G1C7 and G3C3, and one for G3C9 since fragments located in different regions of the 16S rRNA gene were obtained. This analysis confirmed the results obtained previously by Blastn (Fig. 5). Table 4. Autoaggregation and coaggregation capacity of 08 bacterial isolates.
Fig. 4. Agarose gel electrophoresis of PCR products targeting the 16S rRNA gene amplified from the genomic DNA of G1C7, G3C3, G3C7, G3C9, and G3C10. M is a 100 bp molecular weight marker. DiscussionThis investigation focused on the isolation of microorganisms from guinea pig feces, leveraging the transit of fecal pellets through the colon to capture microorganisms from the internal intestinal walls. The utilization of a selective MRS medium for bacterial isolation favored the growth of lactic acid bacteria, contributing to the observed prevalence of Gram-positive bacteria. Furthermore, the coexistence of both Gram-positive and Gram-negative bacteria aligns with the established distribution of predominant phyla in the guinea pig intestine. Specifically, Firmicutes encompasses a substantial population of Gram-positive bacteria, while Bacteroidetes comprises Gram-negative bacterial entities (Crowley et al., 2017). As evidenced by the results, equivalent levels of cocci and bacilli were discerned, accompanied by an increased prevalence of catalase-positive bacteria. This observation diverges from the findings of Serrano et al. (2020), who documented a prevalence of bacillary strains isolated from guinea pig feces, with over 80% of bacteria exhibiting catalase negativity. The criteria for the selection of potential probiotic microorganisms encompassed attributes such as lactic acid bacteria, Gram-positive staining, and catalase negativity (Jung et al., 2020; Castillo et al., 2022). Protease activity emerges as a noteworthy characteristic among probiotic microorganisms, contributing to the generation of bioactive peptides endowed with antimicrobial, immunomodulatory, and antioxidant properties conducive to host well-being (Wang et al., 2021). Notably, all assessed bacteria manifested proteolytic activity, aligning with the findings of Serrano et al. (2020), who identified a substantial proportion of lactic acid bacteria isolated from guinea pig feces exhibiting protease activity, with a heightened prevalence in the Peruvian breed in comparison to the Andean or Inti breed. Furthermore, the levels of proteolysis closely paralleled those observed in probiotics isolated from other species, such as cattle, where indices of proteolytic activity were analogous (0.2–0.6) (Ramadhan et al., 2021). Conversely, an inquiry into probiotics isolated from pig feces revealed proteolytic halos with a maximum diameter of 3 mm following 24 hours of incubation (Marchwińska and Gwiazdowska, 2022). Another investigation focused on probiotics obtained from sheep dairy products reported protein hydrolysis diameters exceeding 10 mm (Silva et al., 2019). The microbial capacity for carbohydrate hydrolysis through the enzymatic actions of amylase and cellulase plays a pivotal role in enhancing food digestibility and optimizing the utilization of the provided diet (Yi et al., 2020). The observed low incidence of bacteria exhibiting amylolytic capacity in this study aligns with previous investigations, revealing only 13% of amylolytic strains isolated from porcine intestines (Marchwińska and Gwiazdowska, 2022), and a mere 8% of strains with recognized probiotic potential isolated from guinea pig feces (Serrano et al., 2020). Notably, the amylolytic levels recorded fall within the spectrum of those reported for amylolytic isolates originating from the intestines of Osphronemus goramy fish (Suprayudi et al., 2016). Table 5. Accession numbers and strain names stored in GenBank with the highest percentage of identity to potential probiotic isolates.
Fig. 5. Phylogenetic trees based on 16S rRNA gene sequences for 05 isolates enclosed in a black rectangle. A) Phylogenetic tree for isolates G3C10 and G3C7. B) Phylogenetic tree for isolates G1C7 and G3C3. C) Phylogenetic tree for isolate G3C9. Conversely, no bacteria with cellulase activity were documented in contrast to other investigations in guinea pigs, where 4% of bacteria exhibited cellulolytic activity (Serrano et al., 2020). Comparable cellulolytic activity has been observed in isolates from the intestinal tract of chickens (Nurliana et al., 2022). Within the context of probiotic attributes, one of the paramount considerations is the resilience of lactic acid bacteria to low pH conditions, with some strains demonstrating tolerance to exceedingly low pH values (pH 2.0). These bacteria exhibit resilience for a minimum of 2 hours, yet optimal growth is observed at pH levels proximate to neutrality (Jomehzadeh et al., 2020; Marchwińska and Gwiazdowska, 2022). Lactic acid bacteria isolated from guinea pig feces have similarly displayed tolerance to pH levels of 2.0 and 4.0 (Serrano et al., 2020). Consequently, the characteristics of the microorganisms elucidated in this investigation concur with these notable attributes. The antagonistic proficiency against pathogenic microorganisms stands as a pivotal criterion in the discerning selection of probiotics. This investigation substantiates the in vitro antagonistic efficacy against Salmonella sp., a precipitating factor in guinea pig mortality, thereby underscoring the potential of these probiotic strains to mitigate Salmonella sp. infections in guinea pig hosts (Ortiz, 2016; Serrano et al., 2020). Moreover, this antagonistic prowess extends to other pathogens, including Yersinia enterocolitica, Shigella flexneri, E. coli, and S. aureus, as documented in existing literature (Jomehzadeh et al., 2020; Ahmed et al., 2021). The hemolytic capacity assessment of the bacterial isolates serves a dual purpose: first, to categorically exclude strains with potential pathogenicity, aligning with the guidelines stipulated by the Food and Agriculture Organization (FAO) regarding the safety imperative for host organisms in the context of probiotic applications (FAO, 2016; Deidda et al., 2020). Second, the observed halos in this study signify a manifestation of alpha hemolysis on blood agar (Halder et al., 2017). Consistent with these findings, prior investigations have reported analogous hemolytic activity in lactic acid bacteria isolated from the gastrointestinal tracts of poultry and swine (Tuyarum et al., 2021; Makzum et al., 2023). The degradation of blood agar is posited to be instigated by hemolysin, an enzymatic agent capable of lysing host cells to facilitate the assimilation of ferrous compounds, particularly hemoglobin (Chen et al., 2018). It is pertinent to note that such hemolytic attributes are conventionally associated with a multitude of pathogenic microorganisms. The phenomenon of autoaggregation is closely linked to the development of biofilms, facilitating enhanced adhesion and colonization within the intestinal environment. Simultaneously, coaggregation serves as a mechanism to impede the adhesion of pathogenic microorganisms to the intestinal epithelium (Venkatasatyanarayana et al., 2017). The observed proficiency in aggregation among the selected isolates aligns with findings reported by Kousha et al. (2021), who assessed the probiotic potential of Lactobacillus strains. This concordance is further substantiated by a study involving Lactobacillus strains isolated from the intestinal tracts of poultry, wherein a majority exhibited autoaggregation ranging between 11% and 29% after a 2 hours evaluation period (Aziz et al., 2019). However, extant literature also documents instances of lactic acid bacteria, sourced from the digestive systems of poultry, as well as dairy and meat products, and some vegetables as fermented Torshi exhibiting higher percentages of autoaggregation and coaggregation against Salmonella sp. within a 2–4 hours assessment window (Reuben et al., 2019; Ahmed et al., 2021; Nemati et al., 2023). Molecular analyses afford heightened precision in the identification of microorganisms possessing optimal probiotic characteristics. The identification of L. citreum, characterized as Gram-positive cocci with negative catalase activity, aligns with reports highlighting its probiotic capacity to inhibit pathogenic microorganisms (Muthusamy et al., 2023; Ahn et al., 2023). Members of this genera also have been isolated from fermented Torshi showing a high acid and bile tolerance (Nemati et al., 2023). This alignment is consistent with the characteristics identified in the present study for isolates G1C7 and G3C3, both classified within the same taxonomic group. Enterococcus gallinarum, identified as Gram-positive cocci with negative catalase activity (Eshaghi et al., 2015), closely mirrors the characteristics of isolate G3C7. This bacterial group is recognized for its probiotic potential, attributed to the production of pathogen-inhibiting bacteriocins, thus conferring them the status of potential immunomodulatory probiotics (Román et al., 2015; Totewad & Gyananath, 2018). Exiguobacterium species, characterized by adaptability to diverse habitats, typically exhibit Gram-positive cocci morphology with catalase activity (Pandey, 2020). It is noteworthy that isolate G3C9, while assigned to this species, manifested an absence of catalase activity. Some members of the Exiguobacterium genus have been reported to harbor probiotic potential associated with nutrient absorption and the reduction of host mortality (Hadi et al., 2014; Cong et al., 2017). Finally, L. lactis, recognized as a Gram-positive cocci with negative catalase activity, holds the status of a probiotic organism (Jung et al., 2020). ConclusionThe five selected isolates manifest commendable attributes qualifying them as potential probiotics. These features encompass protease enzymatic activity, amylase activity in one instance, the ability to inhibit Salmonella sp., resilience to acidic pH conditions, host safety demonstrated by the absence of hemolytic activity, and notable levels of autoaggregation and coaggregation with Salmonella sp. Furthermore, these isolates have been identified as bacteria with antecedent reports detailing their probiotic potential across diverse species. Thus, they could be used to prepare probiotic products to improve the health and productive parameters of guinea pigs. Complement studies should be conducted about potential probiotics of acid lactic isolated from other guinea pig breeds such as Inti and Andina. AcknowledgmentThe authors would like to thank Universidad Nacional Hermilio Valdizan–Peru, for the financing and providing us with the facilities to use the facilities of the Kotosh Production Center to maintain the animals during the experiment. Conflict of interestThe authors declare that there is no conflict of interest. FundingPublication of this research was funded by Universidad Nacional Hermilio Valdizan–Peru, through the resolution N 0244-2022-UNHEVAL-VRI. Author contributionsJGV and MSA participated in the design and conception of the study. MSA, AHB, and WRJ collected the required data. JGV, FAC, and MSA analyzed the collected data and JGV, MSA, EMP, and EM wrote the manuscript. All authors revised and approved the final manuscript. Data availabilityAll data supporting the findings of this study are available within the manuscript. ReferencesAbedi, E. and Hashemi, S.M.B. 2020. Lactic acid production–producing microorganisms and substrates sources-state of art. Heliyon 6(10), e04974. Ahmed, A.S.I., El Moghazy, G.M., Elsayed, T.R., Goda, H.A.L. and Khalafalla, G.M. 2021. Molecular identification and in vitro evaluation of probiotic functional properties of some Egyptian lactic acid bacteria and yeasts. J. Genet. Eng. Biotechnol. 19(1), 1–16. Ahn, H., Lee, G., Lee, W., Kim, M. and Lee, K.G. 2023. Evaluation of probiotic and anti-inflammatory properties of bacteriocinogenic Pediococcus acidilactici HW01 and Leuconostoc citreum HW02 from malted barley. Chem. Biol. Technol. Agric. 10(1), 1–14. Alcívar-Vega, M.D. and Vera-Vargas, V.E. 2013. Aislamiento de bacterias celulolíticas a diferentes profundidades en plantación de Teca (Tectona grandis) y Pechiche (Vitex gigantea), B. S. thesis, CIBESPAM MFL, Manabí, Ecuador. Ampuero-Riega, J. and Morales-Cauti, S. 2021. Determinación de residuos de antibióticos en músculo, hígado y riñón de cuyes comercializados en cuatro ciudades del Perú. Rev. de Investig. Vet. del Peru. 32(1), e19508. Anguiano-Aguilar, E. 2019. Aislamiento, selección y caracterización de bacterias con actividad celulolítica, M. S. tesis, Universidad Michoacana de San Nicolás de Hidalgo, Morelia, Mexico. Angulo-Tisoc, J.M., Jara, L.M., Pacheco, J.I. and Pezo, D. 2021. Frecuencia de agentes bacterianos asociados a mortalidad en cuyes de centros de crianza familiar-comercial en Canchis, Cusco. Rev. de Investig. Vet. del Peru. 32(3), e20415. Aziz, G., Fakhar, H., Ur-Rahman, S., Tariq, M. and Zaidi, A. 2019. An assessment of the aggregation and probiotic characteristics of Lactobacillus species isolated from native (desi) chicken gut. J. Appl. Poult. Res. 28(4), 846–857. Bazán, V., Bezada, S., Carcelén, F., and Yamada, G. 2019. Efecto de la infección subclínica de Salmonella Typhimurium sobre los parámetros productivos en la producción de cuyes de engorde (Cavia porcellus). Rev. Investig. Vet. Peru. 30(4), 1697–1706. Bernatek, M., Żukiewicz-Sobczak, W., Lachowicz-Wiśniewska, S., and Piątek, J. 2022. Factors determining effective probiotic activity: evaluation of survival and antibacterial activity of selected probiotic products using an “In Vitro” study. Nutrients 14(16), 3323. Bhogoju, S. and Nahashon, S. 2022. Recent advances in probiotic application in animal health and nutrition: a review. Agriculture 12(2), 304. Bintsis, T. 2018. Lactic acid bacteria as starter cultures: an update in their metabolism and genetics. AIMS. Microbiol. 4(4), 665–684. Carcelén, F., López, M., San Martín, F., Ara, M., Bezada, S., Ruiz-García, L., Sandoval-Monzón, R., López, S. and Guevara, J. 2021. Effect of probiotics administration at different levels on the productive parameters of guinea pigs for fattening (Cavia porcellus). Open Vet. J. 11(2), 222–227. Castillo, C., Brito, G., Tello, L. and Flores, L. 2022. Biochemical characterization of lactic acid bacteria from the small intestine of piglets as possible probiotic strains. In the ESPOCH Congresses: The Ecuadorian Journal of STEAM, pp: 3–13. Chen, S., Yang, D., Wen, Y., Jiang, Z., Zhang, L., Jiang, J., Chen, Y., Hu, T., Wang, Q., Zhang, Y. and Liu, Q. 2018. Dysregulated hemolysin liberates bacterial outer membrane vesicles for cytosolic lipopolysaccharide sensing. PLoS Pathog. 14(8), e1007240. Cong, M., Jiang, Q., Xu, X., Huang, L., Su, Y. and Yan, Q. 2017. The complete genome sequence of Exiguobacterium arabatum W-01 reveals potential probiotic functions. Microbiol. Open. 6(5), e00496. Crowley, E.J., King, J.M., Wilkinson, T., Worgan, H.J., Huson, K.M., Rose, M.T. and McEwan, N.R. 2017. Comparison of the microbial population in rabbits and guinea pigs by next generation sequencing. PLoS One 12(2), e0165779. Das, T.K., Pradhan, S., Chakrabarti, S., Mondal, K.C. and Ghosh, K. 2022. Current status of probiotic and related health benefits. Appl. Food. Res. 2(1), 100185. Deidda, F., Graziano, T., Amoruso, A., De Prisco, A., Marco, P. and De Prisco, A. 2020. How probiotics may kill harmful bacteria: the in vitro activity against some haemolytic strains. J. Probiotics. Health 8, 216. Enríquez, K.Y. 2019. Evaluación de la calidad de la carne de cuy (Cavia porcellus) suplementada con un simbiótico natural en la etapa de crecimiento, B. S. thesis, UNMSM, Lima, Peru. Eshaghi, A., Shahinas, D., Li, A., Kariyawasam, R., Banh, P., Desjardins, M., Melano, R.G. and Patel, S.N. 2015. Characterization of an Enterococcus gallinarum isolate carrying a dual vanA and vanB cassette. J. Clin. Microbiol. 53(7), 2225–2229. Esser, D., Lange, J., Marinos, G., Sieber, M., Best, L., Prasse, D., Bathia, J., Rühlemann, M.C., Boersch, K., Jaspers, C. and Sommer, F. 2019. Functions of the microbiota for the physiology of animal metaorganisms. J. Innate. Immun. 11(5), 393–404. Fijan, S. 2014. Microorganisms with claimed probiotic properties: an overview of recent literature. Int. J. Environ. Res. Public. Health 11, 4745–4767. Fitriyanto, N.A., Hadi, S., Bahtiyar, M.I., Prasetyo, R.A. and Erwanto, Y. 2020. Characterization and growth profile of proteolytic strain PK-4 isolated from local slaughterhouse wastewater. In BIO Web of Conferences pp: 03001. FAO. 2016. In Probiotics in animal nutrition—production, impact and regulation. Eds., Bajagai, Y.S., Klieve, A.V.,Dart PJ. and Bryden WL,. Rome, Italy: FAO Animal Production and Health,Vol. 179. Frias, H., Valderrama, N.L.M., Flores, G.J., Cornejo, V.G., Del Solar, J.C., Romani, A.C., Bardales, W., Segura, G.T., Polveiro, R.C., Vieira D.D.S., Lopez-Lapa, R.M. and Quintana, J.L.M. 2023. An analysis of the cecum microbiome of three breeds of the guinea pig: Andina, inti, and Peru. Res. Vet. Sci. 161, 50–61. Hadi, J.A., Gutierrez, N., Alfaro, A.C. and Roberts, R.D. 2014. Use of probiotic bacteria to improve growth and survivability of farmed New Zealand abalone (Haliotis iris). N. Z. J. Mar. Freshwater Res. 48(3), 405–415. Halder, D., Mandal, M., Chatterjee, S.S., Pal, N.K. and Mandal, S. 2017. Indigenous probiotic Lactobacillus isolates presenting antibiotic like activity against human pathogenic bacteria. Biomedicines 5(2), 31. Herrera, E., Petrusan, J.I., Salvá-Ruiz, B., Novak, A., Cavalcanti, K., Aguilar, V., Heinz, V. and Smetana, S. 2022. Meat quality of guinea pig (Cavia porcellus) fed with black soldier fly larvae meal (Hermetia illucens) as a protein source. Sustainability 14(3), 1292. Hugo, F.T. 2023. Caracterización del microbioma del ciego en cuyes (Cavia porcellus) de las razas Inti, Perú y Andina, Chachapoyas-2021. Ph.D. dissertation, UNTMR, Chachapoyas, Peru. Jomehzadeh, N., Javaherizadeh, H., Amin, M., Saki, M., Al-Ouqaili, M.T., Hamidi, H., Seyedmahmoudi, M. and Gorjian, Z. 2020. Isolation and identification of potential probiotic Lactobacillus species from feces of infants in southwest Iran. Int. J. Infect. Dis. 96, 524–530. Jain, A., Jain, R. and Jain, S. 2020. Staining methods—simple staining, negative staining, gram’s staining and acid-fast staining. In: basic techniques in biochemistry, microbiology and molecular biology. Springer protocols handbooks. New York, NY: Humana, pp: 111–116. Jung, M.Y., Lee, C., Seo, M.J., Roh, S.W. and Lee, S.H. 2020. Characterization of a potential probiotic bacterium Lactococcus raffinolactis WiKim0068 isolated from fermented vegetable using genomic and in vitro analyses. BMC Microbiol. 20(1), 1–10. Kousha, S., Ahari, H., Karim, G. and Anvar, S.A.A. 2022. Identification of lactobacilli from milk enzymatic clots and evaluation of their probiotic and antimicrobial properties. Food Sci. Technol. 42, e107721. Lord, E., Collins, C., deFrance, S., LeFebvre, M.J., Pigière, F., Eeckhout, P., Erauw, E., Fitzpatrick, S.M., Healy, P.F., Martínez-Polanco, M.F., Garcia, J.L., Ramos-Roca, E., Delgado, M., Sáncehz-Urriago, A., Peña-León, G.A., Toyne, J.M., Dahlstedt, A., Moore, K.M., Laguer-Diaz, C., Zori, C. and Matisoo-Smith, E. 2020. Ancient DNA of guinea pigs (Cavia spp.) indicates a probable new center of domestication and pathways of global distribution. Sci. Rep. 10(1), 8901. Makzum, S., Ghadam, P. and Ramezani, M. 2023. Isolation, functional evaluation of probiotic properties and molecular identification of strains isolated from Iranian poultry’s gut. Iran. J. Microbiol. 15(2), 267–277. Marchwińska, K. and Gwiazdowska, D. 2022. Isolation and probiotic potential of lactic acid bacteria from swine feces for feed additive composition. Arch. Microbiol. 204(1), 61. Mendonça, A.A., Pinto-Neto, W.D.P., da Paixão, G.A., Santos, D.D.S., De Morais Jr, M.A. and De Souza, R.B. 2022. Journey of the probiotic bacteria: survival of the fittest. Microorganisms 11(1), 95. Moon, S.H., Kim, C.R. and Chang, H.C. 2018. Heterofermentative lactic acid bacteria as a starter culture to control kimchi fermentation. Lwt 88, 181–188. Morales, S. 2017. Patógenos bacterianos y parasitarios más frecuentes en cuyes de crianza familiar—comercial en tres distritos de la Provincia de Bolognesi, Departamento de Ancash en época de seca. M. S. thesis, UNMSM, Lima, Peru. Muthusamy, K., Han, H.S., Soundharrajan, I., Jung, J.S., Valan Arasu, M. and Choi, K.C. 2023. A novel strain of probiotic Leuconostoc citreum inhibits infection-causing bacterial pathogens. Microorganisms 11(2), 469. Nemati, V., Hashempour-baltork, F., Alizadeh, A.M. and Varzakas, T. 2023. Production of traditional torba yogurt using lactic acid bacteria isolated from fermented vegetables: microbiological, physicochemical and sensory properties. J. Agric. Food Res. 14, 100850. Nurliana, N., Siregar, B.H., Sari, W.E., Helmi, T.Z. and Sugito, S. 2022. Identification of cellulolytic lactic acid bacteria from the intestines of laying hens given AKBISprob based on 16S ribosomal ribonucleic acid gene analysis. Vet. World. 15(7), 1650–1656. Obregón, R., Serrano-Martínez, E. and Chauca, L. 2018. Causas de mortalidad neonatal en cobayos (Cavia porcellus) durante la estación fría en el Instituto Nacional de Innovación Agraria, Lima-Perú. Salud Tecnol. Vet. 2, 93–99. Oliphant, K. and Allen-Vercoe, E. 2019. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome 7(1), 1–15. Ortiz, J.L. 2016. Lactobacillus sp. como aditivo sobre parámetros productivos en cuy (Cavia porcellus). B. S. thesis, URP, Lima, Peru. Pachla, A., Wicha, M., Ptaszyńska, A.A., Borsuk, G., –Trokenheim, Ł.Ł. and Małek, W. 2018. The molecular and phenotypic characterization of fructophilic lactic acid bacteria isolated from the guts of Apis mellifera L. derived from a polish apiary. J. Appl. Genet. 59, 503–514. Palakawong Na Ayudthaya, S., van der Oost, H., van der Oost, J., van Vliet, D.M. and Plugge, C.M. 2019. Microbial diversity and organic acid production of guinea pig faecal samples. Curr. Microbiol. 76, 425–434. Pandey, N. 2020. Exiguobacterium. In Beneficial microbes in Agro-Ecology. Eds., Amaresan, N., Senthil Kumar, M., Annapurna, K., Kumar, K., Sankaranarayanan, A. Academic Press, pp: 169–183. Plaza-Diaz, J., Ruiz-Ojeda, F.J., Gil-Campos, M. and Gil, A. 2019. Mechanisms of action of probiotics. Adv. Nutr. 11(4), 1054. Quesada, A., Reginatto, G.A., Colantonio, L.D. and Burrone, M.S. 2016. Antimicrobial resistance of Salmonella spp isolated animal food for human consumption. Rev. Peru. Med. 33(1), 32–44. Ramadhan, A.R., Bachruddin, Z., Erwanto, Y. and Hanim, C. 2021. Isolation and selection of proteolytic lactic acid bacteria from colostrum of dairy cattle. In IOP Conference Series: Earth and Environmental Science. 788(1), 012077. Reuben, R.C., Roy, P.C., Sarkar, S.L., Alam, R.U. and Jahid, I.K. 2019. Isolation, characterization, and assessment of lactic acid bacteria toward their selection as poultry probiotics. BMC Microbiol. 19, 1–20. Reynaga-Rojas, M.F., Vergara-Rubín, V., Chauca-Francia, L., Muscari-Greco, J. and Higaonna-Oshiro, R. 2020. Sistemas de alimentación mixta e integral en la etapa de crecimiento de cuyes (Cavia porcellus) de las razas Perú, Andina e Inti. Rev. investig. vet. Peru. 31(3), e18173. Román, L., Padilla, D., Acosta, F., Sorroza, L., Fátima, E., Déniz, S., Grasso, V., Bravo, J. and Real, F. 2015. The effect of probiotic Enterococcus gallinarum L-1 on the innate immune parameters of outstanding species to marine aquaculture. J. Appl. Anim. Res. 43(2), 177–183. Saldarriaga-Barrón, M.F. 2018. Efecto del uso de probióticos en cuyes (Cavia porcellus) de engorde desafiados con Salmonella Typhimurium sobre los parámetros productivos y sanguíneos. B. S. thesis. UNMSM, Lima. Peru. Serrano, C., Jara, L.M., Chauca, L. and Shiva, C. 2020. Evaluación In Vitro de la capacidad probiótica de bacterias ácido-lácticas aisladas de heces de cuyes (Cavia porcellus) de un centro experimental. Salud Tecnol. Vet. 8(2), 40–46. Silva, E.O.O., Nespolo, C.R., Sehn, C.P., Pinheiro, F.C. and Stefani, L.M. 2019. Lactic acid bacteria with antimicrobial, proteolytic and lipolytic activities isolated from ovine dairy products. Food Sci. Technol. 40, 293–299. Solden, L.M., Naas, A.E., Roux, S., Daly, R.A., Collins, W.B., Nicora, C.D., Purvine, S.O., Hoyt, D.W., Schückel, J., Jørgensen, B., Willats, W., Spalinger, D.E., Firkins, J.L., Lipton, M.S., Sullivan, M.B., Pope, P.B. and Wrighton, K.C. 2018. Interspecies cross-feeding orchestrates carbon degradation in the rumen ecosystem. Nat. Microbiol. 3(11), 1274–1284. Suprayudi, M.A., Zairin Jr, M. and Sunarno, M.T.D. 2016. Screening of probiotics from the digestive tract of gouramy (Osphronemus goramy) and their potency to enhance the growth of tilapia (Oreochromis niloticus). Aquac. Aquar. Conserv. Legis. 9(5), 1121–1132. Tamura, K., Stecher, G. and Kumar, S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38(7), 3022–3027. Tang, C., Ma, J., Kong, F., Li, B., Du, Q., Zhang, Y., Wang, H., Tang, Q., Hu, S., Liu, L., Li, X. and Li, M. 2022. The analysis of transcriptomes and microorganisms reveals differences between the intestinal segments of guinea pigs. Animals 12(21), 2925. Totewad, N. and Gyananath, G. 2018. Identification of bacteriocin producing Enterococcus Gallinarum N3 from intestine of freshwater fish Cyprinus Carpio. J. Emerg. Technol. Innov. Res. 5(10), 255–263. Tuo, Y., Yu, H., Ai, L., Wu, Z., Guo, B. and Chen, W. 2013. Aggregation and adhesion properties of 22 Lactobacillus strains. J. Dairy. Sci. 96(7), 4252–4257. Tuyarum, C., Songsang, A. and Lertworapreecha, M. 2021. In vitro evaluation of the probiotic potential of Lactobacillus isolated from native swine manure. Vet. World. 14(5), 1133. Venkatasatyanarayana, N., Vishwanathan, S. and Kadirvelu, J. 2017. Molecular characterization of antimicrobial Lactobacillus isolates and evaluation of their probiotic characteristics in vitro for use in poultry. Food Biotechnol. 31(1), 20–41. Wang, H., Zhang, X., Chen, Z., Hao, G. and Li, G. 2021. Two potential probiotic bacillus with proteolytic activity to dietary protein from adult feces. Biocontrol. Sci. 26(4), 221–224. Wang, Y., Wu, J., Lv, M., Shao, Z., Hungwe, M., Wang, J., Bai, X., Xie, J., Wang, Y. and Geng, W. 2021. Metabolism characteristics of lactic acid bacteria and the expanding applications in food industry. Front. Bioeng. Biotechnol. 9, 612285. Yi, R., Pan, Y., Long, X., Tan, F. and Zhao, X. 2020. Enzyme producing activity of probiotics and preparation of compound enzyme. J. Chem. 2020, 1–8. Zheng, D., Liwinski, T. and Elinav, E. 2020. Interaction between microbiota and immunity in health and disease. Cell Res. 30(6), 492–506. | ||
| How to Cite this Article |
| Pubmed Style Goicochea-vargas J, Salvatierra-alor M, Acosta-pachorro F, Rondón-jorge W, Herrera-briceño A, Morales-parra E, Mialhe E. Genomic characterization and probiotic potential of lactic acid bacteria isolated from feces of guinea pig (Cavia porcellus). Open Vet. J.. 2024; 14(2): 716-729. doi:10.5455/OVJ.2024.v14.i2.12 Web Style Goicochea-vargas J, Salvatierra-alor M, Acosta-pachorro F, Rondón-jorge W, Herrera-briceño A, Morales-parra E, Mialhe E. Genomic characterization and probiotic potential of lactic acid bacteria isolated from feces of guinea pig (Cavia porcellus). https://www.openveterinaryjournal.com/?mno=180402 [Access: January 24, 2026]. doi:10.5455/OVJ.2024.v14.i2.12 AMA (American Medical Association) Style Goicochea-vargas J, Salvatierra-alor M, Acosta-pachorro F, Rondón-jorge W, Herrera-briceño A, Morales-parra E, Mialhe E. Genomic characterization and probiotic potential of lactic acid bacteria isolated from feces of guinea pig (Cavia porcellus). Open Vet. J.. 2024; 14(2): 716-729. doi:10.5455/OVJ.2024.v14.i2.12 Vancouver/ICMJE Style Goicochea-vargas J, Salvatierra-alor M, Acosta-pachorro F, Rondón-jorge W, Herrera-briceño A, Morales-parra E, Mialhe E. Genomic characterization and probiotic potential of lactic acid bacteria isolated from feces of guinea pig (Cavia porcellus). Open Vet. J.. (2024), [cited January 24, 2026]; 14(2): 716-729. doi:10.5455/OVJ.2024.v14.i2.12 Harvard Style Goicochea-vargas, J., Salvatierra-alor, . M., Acosta-pachorro, . F., Rondón-jorge, . W., Herrera-briceño, . A., Morales-parra, . E. & Mialhe, . E. (2024) Genomic characterization and probiotic potential of lactic acid bacteria isolated from feces of guinea pig (Cavia porcellus). Open Vet. J., 14 (2), 716-729. doi:10.5455/OVJ.2024.v14.i2.12 Turabian Style Goicochea-vargas, José, Max Salvatierra-alor, Fidel Acosta-pachorro, Wilson Rondón-jorge, Arnold Herrera-briceño, Edson Morales-parra, and Eric Mialhe. 2024. Genomic characterization and probiotic potential of lactic acid bacteria isolated from feces of guinea pig (Cavia porcellus). Open Veterinary Journal, 14 (2), 716-729. doi:10.5455/OVJ.2024.v14.i2.12 Chicago Style Goicochea-vargas, José, Max Salvatierra-alor, Fidel Acosta-pachorro, Wilson Rondón-jorge, Arnold Herrera-briceño, Edson Morales-parra, and Eric Mialhe. "Genomic characterization and probiotic potential of lactic acid bacteria isolated from feces of guinea pig (Cavia porcellus)." Open Veterinary Journal 14 (2024), 716-729. doi:10.5455/OVJ.2024.v14.i2.12 MLA (The Modern Language Association) Style Goicochea-vargas, José, Max Salvatierra-alor, Fidel Acosta-pachorro, Wilson Rondón-jorge, Arnold Herrera-briceño, Edson Morales-parra, and Eric Mialhe. "Genomic characterization and probiotic potential of lactic acid bacteria isolated from feces of guinea pig (Cavia porcellus)." Open Veterinary Journal 14.2 (2024), 716-729. Print. doi:10.5455/OVJ.2024.v14.i2.12 APA (American Psychological Association) Style Goicochea-vargas, J., Salvatierra-alor, . M., Acosta-pachorro, . F., Rondón-jorge, . W., Herrera-briceño, . A., Morales-parra, . E. & Mialhe, . E. (2024) Genomic characterization and probiotic potential of lactic acid bacteria isolated from feces of guinea pig (Cavia porcellus). Open Veterinary Journal, 14 (2), 716-729. doi:10.5455/OVJ.2024.v14.i2.12 |