| Research Article | ||
Open Vet. J.. 2024; 14(5): 1117-1129 Open Veterinary Journal, (2024), Vol. 14(5): 1117–1129 Research Article Occurrence, antimicrobial resistance, and molecular characterization of Salmonella enterica from chicken products and human in Wasit Governorate of IraqAhmed Abdali Jabor Al-Shafee* and Mushtaq Talib AbdulwahidDepartment of Public Health, College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq *Corresponding Author: Ahmed Abdali Jabor Al-Shafee. Department of Public Health, College of Veterinary Medicine, University of Baghdad, Baghdad, Iraq. Email: Ahmed.Abd1104e [at] covm.uobaghdad.edu.iq Submitted: 19/01/2024 Accepted: 07/04/2024 Published: 31/05/2024 © 2024 Open Veterinary Journal
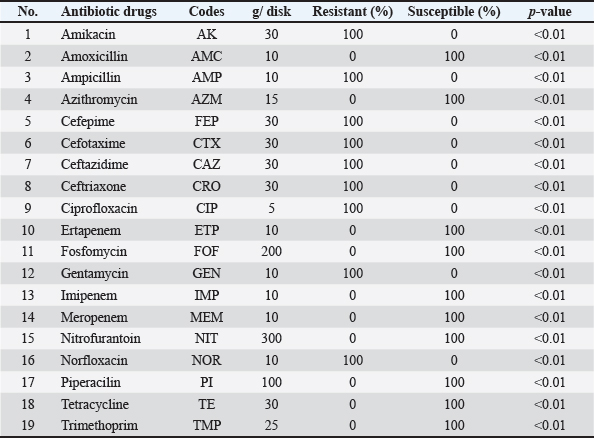
AbstractBackground: Salmonella infections are considered the most common foodborne pathogens responsible for zoonotic infections and food poisoning in humans and animal species such as birds. Antimicrobial resistance is considered a global anxiety because it causes human public health repercussions, as well as leads to an increase in animal morbidity and death. Aim: The aims of this study are the isolation and identification of Salmonella enterica, as well as to investigate the antimicrobial susceptibility test (AST) and the molecular characteristics using polymerase chain reaction (PCR) and sequences for isolates from chicken products (eggs, livers, and minced meat) and human in the Wasit Governorate of Iraq. Methods: A total of 300 samples (150 chicken product samples including eggs, livers, and minced meat, and 150 human fecal samples) were collected from the Wasit governorate of Iraq from January to December 2022. The bacterial isolation was done according to recommendations of ISO 6579 standard and the Global Foodborne Infections Network laboratory protocol. Serotyping test and AST were done by using 19 antibiotic agents according to the recommendations of the Clinical and Laboratory Standards Institute, 2022 by using disc diffusion susceptibility test and Vitik 2 test. Finally, the suspected isolates were confirmed using the conventional PCR method and sequencing for a unique rRNA gene. Results: The results showed that the isolation percentage of S. enterica in chicken products was 8.66% (12% eggs, 6% livers, and 8% minced meat), while in humans it was 4.6%. Also, showed 100% of Salmonella typhi in humans. While, in chicken eggs S. typhi, Salmonella typhimurium, and Salmonella enteritidis were 50%, 33.33%, and 16.66%, respectively. Also, showed 100% of S. typhimurium in both livers and minced meat. The AST in human isolates showed resistance to Ampicillin, Cefotaxime, Ceftazidime, Cefepime, Amikacin, Gentamicin, Ciprofloxacin, Norfloxacin, and Ceftriaxone, while no resistance to Amoxicillin, Pipracillin, Ertapenem, Imipenem, Meropenem, Fosfomycin, Nitrofurantoin, Trimethoprim, Azithromycin, and Tetracycline. In chicken products, isolates were resistant with different percentages to Amikacin, Gentamicin, Tetracycline, Ciprofloxacin, Norfloxacin, Nitrofurantoin, Ampicillin, Cefotaxime, Ceftazidime, Cefepime, and Trimethoprim; while no resistance to Amoxicillin, Pipracillin, Ertapenem, Imipenem, Meropenem, Fosfomycin, Azithromycin, and Ceftriaxone. Sequencing by using rRNA gene was done for four PCR products. Conclusion: This study showed the presence of genetic mutations for S. enterica which led to variations in the molecular characteristics, and antimicrobial drug resistance of S. enterica isolated from chicken products and humans. Keyword: Salmonella enterica, Chicken eggs, Antimicrobial susceptibility test, Molecular characterization, PCR. IntroductionSalmonella enterica is considered the most common foodborne pathogen which is responsible for food poisoning and zoonotic infections in humans and animal species such as birds (Jajere, 2019). More than 2,600 serovars of this bacterium which mostly can be transmitted through food contamination to humans, especially by those that come from animal origin, and in particular chicken products (eggs, livers, and minced meat), and other kinds of meat (Chaudhary et al., 2022). These bacteria characterized by rod-shaped, Gram-negative, facultative anaerobes, motile, belong to the family of Enterobacteriaceae (Crump et al., 2015; Kanaan, 2023). Many disease syndromes are caused by different serovars of S. enterica ranging from Typhoid fever (Enteric or Systemic) which is caused by Salmonella typhi and Salmonella paratyphi A; to gastroenteritis (Intestinal or Diarrheal) caused by non-typhoidal Salmonella (NTS) (Salmonella typhimurium and Salmonella enteritidis); and the severity of infection was varies depending on the type of the host (Feasey et al., 2012; Jaslow et al., 2018; Hanoun and Al-Samrraae, 2019). Each year about 37% of typhoidal Salmonella and 52% of NTS cases; also, approximately, 1.3 billion cases, with three million deaths occur yearly (CDC, 2015). There are many diseases significant to poultry; Fowl typhoid caused by S.enterica serovar Gallinarum; Pullorum disease caused by S. enterica serovar Pullorum; and Paratyphoid disease caused by S. typhimurium, S. enteritidis, and S. enterica subsp. Arizonae (Al-Juburi and AL-Sammarraae, 2023). Chickens are the most prevalent type of fowl around the world. Due to its lower amounts of cholesterol and saturated fat than meat from other animals, chicken is usually seen as being healthier than red meat (Abdulwahid et al., 2017; Sahib et al., 2021). Many virulence factors of Salmonella spp. play important roles in the pathogenesis of this bacterium that is used in invading, attaching, and by passing the host's intestinal defense mechanisms. These factors included biofilm, flagella, capsule, plasmids, adhesion systems, and type III secretion systems encoded on the Salmonella pathogenicity island (SPI), types 1 and 2, and other SPIs. The increase in the pathogenicity of microorganisms lead to increase in the resistance of antimicrobial drugs. Antimicrobial resistance (AMR) in human and animal considered a global anxiety, because it causes human public health repercussions, as well as lead to increase in animal morbidity and death. The multi-drug resistance (MDR) bacteria causes a threats to human and animal health and food safety (Xu et al., 2020). AMR can result from a variety of mechanisms, such as: modifications to the target sites, permeability of bacterial cell walls, enzymatic drug modifications, biofilm formation and the energy-dependent clearance of antimicrobials by membrane-bound efflux pumps. Antimicrobial drug resistance indicates that the medication is ineffective for treating clinical disease brought on by a certain bacterial pathogen. Salmonella spp. is one of the most antimicrobial-resistant bacteria, and it is relatively simple for these organisms to develop resistance to new antibacterial medications due to genetic alterations that result in the failure of treatments for its infections in both humans and animals. Although misuse and excessive use of antibiotics result in resistant germs (Abraham et al., 2016). According to Al-Kassie (2009) and (Al-Khikani, 2020), the usage of Antibiotics in human and poultry is important as the following:
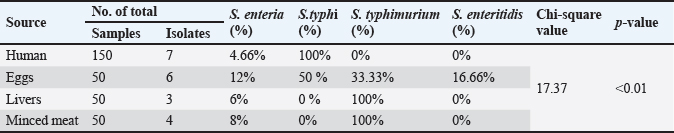
The effects of antibiotics on the intestinal microbiota and host physiology are by: preventing pathogen colonization, having an impact on the immune system, increasing fat absorption by reducing the hydrolysis of conjugated bile salts, and altering the intestinal wall and lamina propria. Overusing and abusing antimicrobial drugs hasten the development of the natural process. A section of the microbiome's DNA that controls susceptibility to a particular antibacterial causes a spontaneous mutation, which leads to the development of antibiotic resistance. Also, the resistance may occur due to transmission of genetic material across bacteria or may occur due to a variety of mechanisms, such as modifications to the target sites or permeability of bacterial cell walls, enzymatic drug modifications, biofilm formation, and the energy-dependent clearance of antimicrobials by membrane-bound efflux pumps (Cuong et al., 2020). There are many bacteria that are resistant to different antibiotics. Antibiotic resistance can be acquired or mutated suggesting that the alterations in the bacteria that prevent the antibiotics from reaching and damaging it are the target (Ansiliero et al., 2021). The antibiotic residues are molecules that persist in the flesh and organs of animals that have been slaughtered after receiving antibiotics without following the drug's withdrawal period. Due to their widespread availability and low cost, antibiotics have been utilized extensively in animal husbandry worldwide. As a result, chickens are a concern for human health due to their detrimental effects on consumer welfare. Additional research has demonstrated that continuous exposure to high levels of antimicrobials derived from animal sources may exacerbate immune responses in people with compromised immune systems and negatively affect the intestinal gut microbiome (Jammoul and El-Darra, 2019). The liver and kidney typically have larger levels of medication residues than the muscles. Consumer health may suffer significantly as a result of the presence of antibiotic residues in food of animal origin, noncompliance with the dosage and withdrawal time recommendations, and poor livestock production methods. Consuming antibiotic residues through animal products may cause the development of antimicrobial drug resistance, hypersensitivity reaction, cancer, mutagenicity, nephropathy, hepatotoxicity, reproductive disorders, bone marrow toxicity, disruption of normal intestinal flora and allergy (Stella et al., 2020; Karasu and Öztürk, 2021; Arsène et al., 2022). Materials and MethodsCollection of samplesA total of 300 samples (150 chicken product samples including 50 eggs, 50 livers, and 50 minced meat from different local supermarkets, and 150 human fecal samples from different clinical cases of different Hospitals) were collected from Wasit Governorate of Iraq from January to December 2022, for the isolation and studying the molecular characteristics and AMR of S. enterica. The samples were collected in a sterile condition, then labeled with the information of the clinical cases, and by the enteric transport media (ETM® R060450) the samples were transported by cooler boxes to the microbiology laboratory of Al-Suwaira General Hospital for culturing. The isolation and identification of Salmonella spp. from chicken product samples were performed according to recommendations by the ISO 6579 standard (ISO 6579, 2012). Each sample was weighed 25 g and mixed in a sterile flask with 225 ml of buffered peptone water, then incubated overnight at 37°C for 18–24 hours, then, 1 ml of the suspension was added to 9 ml of Tetrathionate broth (Oxoid-England), followed by incubation for 18–24 hours at 42°C. After that, culturing by streaking on conventional media (MacConkey and blood agars) (Oxoid-England), and culturing on selective media (Salmonella- Shigella Agar (SS agar) (Condalab-Spain), Xylose Lysine Deoxycholate agar (XLD) (Oxoid-England), Hektoen Enteric Agar (HEA) (Liofilchem-Italy), and Chromogenic medium (Chrom-French)). Then incubation of plates at 37°C for 18–24 hours. After that molecular identification polymerase chain reaction (PCR) and sequencing of the isolates were done. The isolation and identification of Salmonella spp. from human stool specimens were performed according to the recommendation of the Global Foodborne Infections Network laboratory protocol, as the following: - Suspended 1 g of feces in 9 ml of buffered peptone water and incubated for 18–24 hours. at 37°C, then, 1 ml of the suspension was added to 9 ml of Tetrathionate broth and incubated for 18–24 hours, at 42°C. For confirmation of the diagnosis, the blood samples were taken from the same patients with positive results. The culturing, diagnosis of the samples, serotyping, molecular identification, and sequencing of the isolates were done by the same steps mentioned above in chicken product samples. Biochemical test was done by using the Analytical Profile Index 20E (API 20E) and Vitik 2-test. Serotyping test and conventional PCR for Salmonella spp. isolates were performed in the Central Laboratory of the Iraqi Ministry of Health as described by Kipper et al. (2019). The antimicrobial susceptibility test (AST) was done by disc diffusion susceptibility test and Vitik 2 test, using 19 antibiotic agents according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI, 2022). Molecular detectionThe technique PCR was used to detect S. enterica from environmental and clinical samples (Nakano et al., 2012). Cycling conditions of PCR were accomplished to adjust the optimum temperature, time, number of cycles, denaturation, annealing, and extension stages. The conventional PCR for all isolates (20 isolates) was done by amplifying the identity rRNA gene of S. enterica (865 bp) using primers (-F 5′– CGATGCGTTGAGCTAACCGG–3′) and (-R 5′– CAGAAGCGATAACCACGTCGTC –3′) that tested in the National Center for Biotechnology Information (NCBI) Genbank data base and used in the current study as described by Kipper et al. (2019). Finally, the sequencing for some isolates of S. enterica was done. Forward and reverse primers provided by Alpha DNA company, Canada were used to amplify specific DNA fragment of rRNA gene. Sequences and phylogeny analysisFour PCR products for the identity rRNA gene of some isolates of S. enterica from clinical human and chicken product samples were stored at −20°C, then these DNA samples along with primer were sent to the Macrogen Company in Korea to perform sequencing of their nucleotides. Sequences were determined by an automatic sequencer (NovaSeq X Plus/Korea). These DNA sequences were analyzed and the similarity was achieved using Basic Local Alignment Search Tool (BLAST) in NCBI. By using the MEGA 11 method, the evolutionary history was inferred and evolutionary analyses were conducted. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. By using the Maximum Composite Likelihood method, the evolutionary distances were detected in the units of the number of base substitutions per site. After PCR amplification, the products were separated on a 1.5% Agarose gel electrophoresis and visualized by exposure to ultra violate light (302 nm) after dyeing by using red safe dye (Nucleic acid staining solution). Sequencing of the gene was performed, and the applied Biosystem and Homology search was conducted using BLAST program of NCBI and BioEdit program. ResultsResults of isolationThe results of the total collected 300 samples (150 samples from chicken products (50 eggs, 50 livers, and 50 minced meat), and 150 fecal human samples) showed that the total percentage of isolation for S. enterica was 8.66% (13/150) in the chicken products where the highest percentage was recorded in eggs (12%) while the lower percentage was recorded in the livers (6%), also recorded (8%) in the minced meat. On the other hand, 4.6% of the isolates were recovered from humans (Table 1). For all isolates of S. enterica, the genomic DNA was successfully extracted, and the bands of DNA were detected on Agarose gel and visualized under a UV transilluminator that showed 100% extracted genomic DNA. The purity and concentration of extracted DNA were determined directly by a Nanodrop device that ranged between 1.8 and 2. The isolation and identification characteristics by cultural examination of S. enterica isolates that including: colony morphology on MacConkey and blood agars, and selective media such as XLD agar, S.S agar, HEA, and Salmonella Chromogenic medium, also, Gram staining, and microscopic examination, along with conventional biochemical reactions were similar to the phenotypic characteristics that described by Markey et al. (2014). The results of API 20E and VITEK® 2 Test were explained according to the instructions of the manufactures' companies. Table 1. Distribution of total S. enterica isolates in human and chicken products samples.
Table 2. Isolation percentages of S. enterica and their serotypes in human and chicken products isolates.
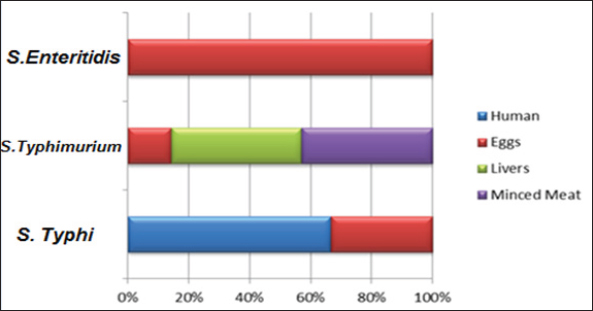
Fig. 1. Percentages of S. enterica serotypes isolated from human and chicken products. Results of serotyping testThe results of the serotyping test showed that the total percentage of S. typhi was 50%, S. typhimurium was 45%, and S. enteritidis was 5% for all isolates of chicken products and human. Also, the results showed 100% (7/7) of S. typhi in humans, while in the eggs were 50% (3/6) of S. typhi, 33.33% (2/6) of S. typhimurium, and 16.66% (1/6) of S. enteritidis. Also, the results showed 100% of S. typhimurium in both livers (3/3) and minced meat (4/4) (Table 2 and Fig. 1). Results of molecular examinationThe results of molecular detection for the identity rRNA gene of S. enterica from all isolates of chicken product and human showed in the Figure 2. Results of sequencing testTo confirm the diagnosis of S. enterica, sequencing was done for four PCR products that were isolated from different sources by amplifying the primer set of the identity rRNA gene for S. enterica serovares (S. typhi, S. typhimurium, and S. enteritidis). All PCR products that sent for sequencing were registrated for the first time on the NCBI under the accession numbers: OQ259883, OQ259884, OQ259885, and OQ259886.
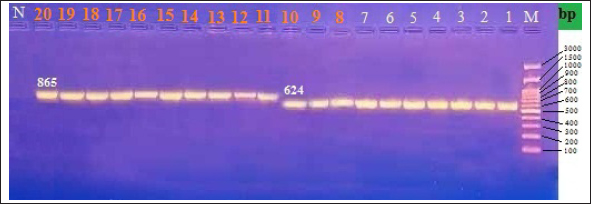
Fig. 2. Gel electrophoresis (2% agarose) of the rRNA gene (865 bp and 624 bp). *Lane M: DNA ladder. *Lane N: negative control. *Lanes (1-7): DNA bands of rRNA gene of S. enterica isolated from human. *Lanes (8-20): DNA bands of rRNA gene of S. enterica isolated from chicken products.
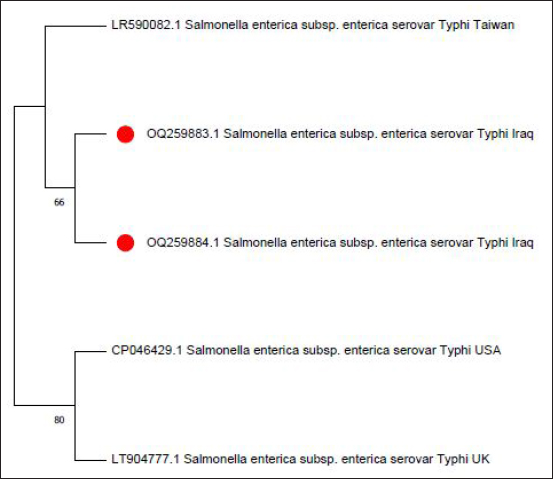
Fig. 3. Phylogenetic tree of sense flanking the rRNA gene of two isolates (OQ259883 and OQ259884) of S. enterica (S. typhi) isolated from Iraq during 2022, compared with the related strains documented in the Genbank. Phylogenetic analysis was conducted using MEGA 11 program. The sequencing and analyzing for similarity by using BLAST in NCBI compare with the global isolates that showed for S. typhi isolates, there were 99% identity for OQ259883 and 99% identity for OQ259884 among global isolates of S. typhi as drown in Phylogenetic tree (Fig. 3). The nucleotide sequencing for these two isolates (OQ259883 and OQ259884) showed two nucleotide substitutions in the isolate OQ259883, which were, Guanine G>A to Adenine and Adenine A> G to Guanine. The isolate OQ259885 (S. typhimurium) showed 98% identity among global isolates as drown in Phylogenetic tree (Fig. 4), and there was no nucleotide substitution.
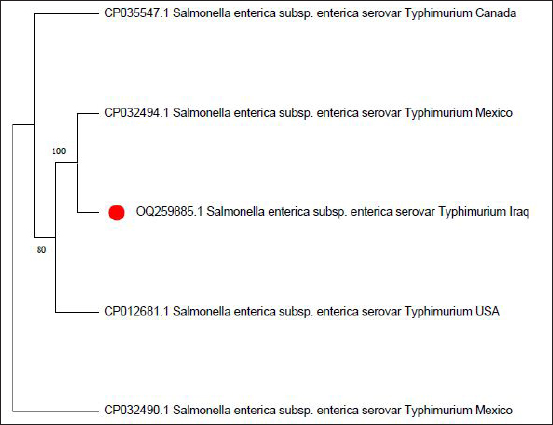
Fig. 4. Phylogenetic tree of sense flanking the rRNA gene of S. enterica (S. typhimurium) (OQ259885) isolated from Iraq during 2022, compared with the related strains documented in the Genbank. Phylogenetic analysis was conducted using MEGA 11 program.
Fig. 5. Phylogenetic tree of sense flanking the rRNA gene of S. enterica (S. enteritidis) (OQ259886) isolated from Iraq during 2022, compared with the related strains documented in the Genbank. Phylogenetic analysis was conducted using MEGA 11 program.
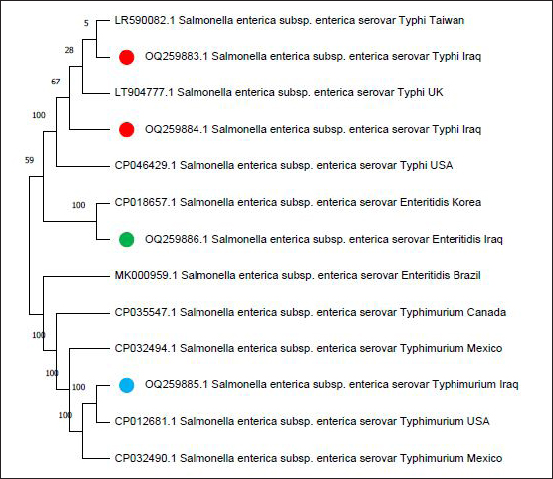
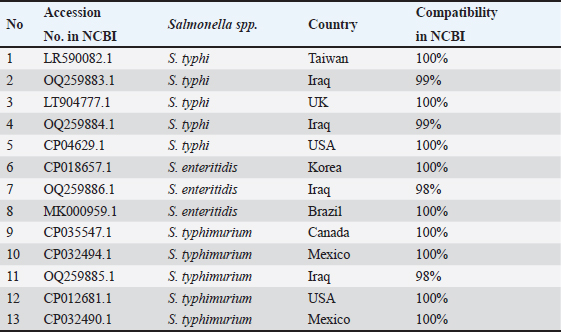
Fig. 6. The phylogenetic tree of sense flanking the rRNA gene of four diagnostic isolates (OQ259883, OQ259884, OQ259885, and OQ259886) with three S. enterica serovars (S. typhi, S. typhimurium, and S. enteritidis) that isolated from Iraq during 2022, and compared with the related all strains documented in the Genbank using MEGA 11 program. Table 3. Sequencing analysis for rRNA gene of S. enterica, in compared with related identity to Genbank strains using BLAST software in NCBI.
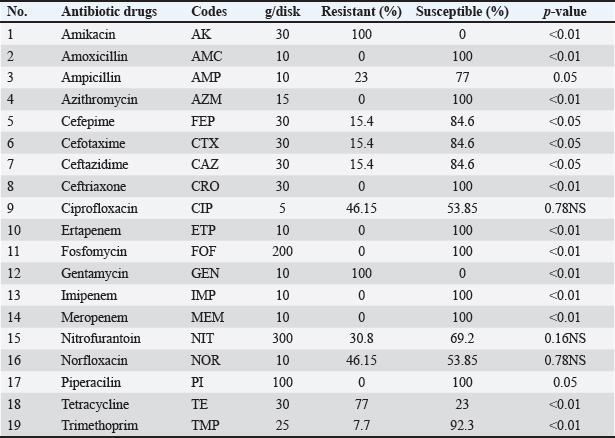
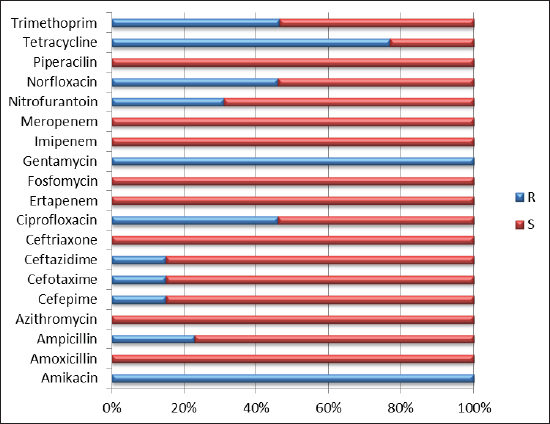
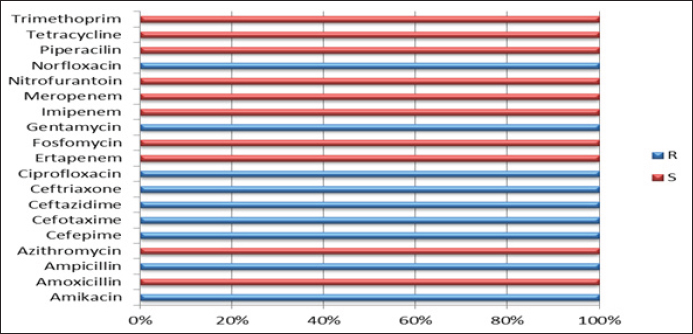
The isolate OQ259886 (S. enteritidis) showed 98% identity among global isolates as drown in Phylogenetic tree (Fig. 5), and there was no nucleotide substitution. The results of evolutionary relationship of taxa S. enterica that were conducted among the Iraqi isolates and the global S. enterica serovars which documented on NCBI database, and that drown of the phylogenetic tree and nucleotide sequencing of sense flanking rRNA gene of four diagnostic isolates (OQ259883, OQ259884, OQ259885, and OQ259886) with three S. enterica serovars (S. typhi, S. typhimurium, and S. enteritidis) that isolated from Iraq during 2022, and compared with the related all strains documented in the Genbank, showed similarities with other nations, including the USA, UK, China, Taiwan, Thailand, and Korea (Fig. 6 and Table 3). Results of ASTThe results of AST showed in the chicken products isolates 100% resistance for Amikacin and Gentamicin; 77% Tetracycline; 46.15% for Ciprofloxacin and Norfloxacin; 30.8% Nitrofurantoin; 23% Ampicillin; 15.4% for Cefotaxime, Ceftazidime, and Cefepime; 7.7% Trimethoprim; while no resistance (0%) for each of Amoxicillin, Pipracillin, Ertapenem, Imipenem, Meropenem, Fosfomycin, Azithromycin, and Ceftriaxone (Table 4 and Fig. 7). Also, we founded that the total percentage of resistance for antibiotic drugs in chicken products isolates were 15.79% (3/19), while the total percentage of susceptibility were 84.21% (16/19). The results of AST in human isolates showed complete resistance 100% for: Ampicillin, Cefotaxime, Ceftazidime, Cefepime, Amikacin, Gentamicin, Ciprofloxacin, Norfloxacin, and Ceftriaxone, while no resistance (0%) for: Amoxicillin, Pipracillin, Ertapenem, Imipenem, Meropenem, Fosfomycin, Nitrofurantoin, Trimethoprim, Azithromycin, and Tetracycline (Table 5 and Fig. 8). Also, we founded that the total percentage of resistance for antibiotic drugs in human isolates were 47.37% (9/19), while the total percentage of susceptibility were 52.63% (10/19). DiscussionBy observation the results of isolation of S. enterica from chicken products and humans, we found that there are significant differences between the serotypes of S. enterica at p < 0.05. The results of this study were agree with (Nader et al., 2015) who isolated S. enterica from raw chickens meat and diarrheic patients as 4% and 10%, respectively, from markets and Al-Yarmmok hospital in Baghdad City of Iraq. Also, this study agree with (Kanaan et al., 2022) who recorded 5% by isolating this bacterium from chicken meat and egg samples in Iraq. Additionally, this study with agreement of (Harb et al., 2019) who reported that 26% of fresh retail chicken meat samples and 39% of raw chicken carcasses were contaminated with Salmonella spp. In Iraq, the results of this study were lower than the results of (Jaffer, 2013) who isolated S. enterica in Iraq from chicken eggs and recorded 30%. Also were lower than the results of (Siddique et al., 2021) who isolated S. enterica from poultry and its associated food products in Pakistan, that recorded 25.67%. The results of this study considered higher than (Rahmani et al., 2011) who isolate Salmonella spp. in Iran from birds and recorded 2.8%. The variance in the percentages of high prevalence of S. enterica in chicken products compare with that in humans may be due to the location and timing of sampling, collection, geographic climate, immunity of human or chickens, age, consumption of drugs, and hygienic restrictions, that may be linked to the relative differences in results between different places. The handling with raw poultry carcasses, consumption of improperly cooked poultry meat, cross-contamination from workers' hands, tools, and utensils, also, handling and contamination with the feces of chickens may be the most frequent sources of Salmonella infection in humans. The high prevalence of Salmonella spp. in the previous studies that comparable to this study could due to the low hygienic standards during the handling in the processes of slaughtering, scalding, de-feathering, evisceration, and carcass cutting. These processes allow for the cross-contamination of healthy and clean birds with diseased ones or contaminated carcasses, and then with human beings. Additionally, the lack of veterinary oversight could result in the slaughter of sick chickens, then spread the infections (Kanaan, 2023). Associations between the detection of S. enterica and different sources of sample with additional factors were examined by using an exact test and a p-value <0.05 considered significant. The results of serotyping test of this study were agree with (Zubair et al., 2017). Who recorded 4.85% of Salmonella spp. isolated from poultry eggs in Duhok/ Iraq, also recorded, 11.75%, 29.4%, and 58.8% for S. typhi, S. typhimurium, and S. enteritidis, respectively. This study with agreement of Kanaan et al. (2022) who isolated S. enteritidis, from chicken meat and egg samples in Iraq, and recorded 5.1% and 4.9% from eggs and chicken meats, respectively. Also, this study agree with (Yousif and Harab, 2011) who isolated Salmonella spp. from children in Thi-Qar governorate of Iraq who recorded 11.17%; but disagree by recorded 42.1% for S. typhimurium and 5.27% for S. enteritidis. Fecal contamination is the main source of transmission for Salmonella spp., the transmission may occur horizontally and vertically. The horizontal transmission occur due to the contamination during the handling in the processes of slaughtering, scalding, de-feathering, evisceration and carcass cutting. Additionally, the infection may occur also through nutrition and water contamination that spread the infection individually or to whole chicken farm, and then to the human beings. In vertical transmission, this bacterium can pass from one generation of poultry to the next via the egg (Hassan et al., 2016). Salmonella spp. spreads through the environment, even after the pathogenic bird has stopped shedding bacteria, the environment can still be infectious for a long time, but it becomes less infectious with time (Thomas et al, 2009). The presence of S. typhi in the chicken eggs, may be due to that the chicken considered as carrier or reservoir by this bacterium that presents inside and then transmitted to the eggs vertically; or may be horizontally by fecal contamination of the chicken eggs during the presence of eggs on the floor, handling of raw poultry carcasses and eggs, cross-contamination from hands of workers by handling with the fecal contamination to the chicken eggs in different stages starting from collecting eggs in the egg production fields, as well as direct contact with eggs in commercial markets, and ending with preparing food inside homes and dealing with preparing egg dishes (Al-Zuhariy, 2022). Table 4. Antibiotic susceptibility test for S. enterica isolated from chicken products.
Fig. 7. Results of antibiotic susceptibility test for S. enterica isolated from chicken products. Table 5. Antibiotic susceptibility test for S. enterica isolated from human.
Fig. 8. Results of antibiotic susceptibility test for S. enterica isolated from human. DNA sequences are crucial for basic biological research as well as for many applied sectors, including biological applications, forensic biology, biotechnology, as well as medical diagnosis. Rapid growth and development in the use of modern DNA sequencing technology plays a significant role in the complete DNA sequencing methods, or the sequencing genomes of many types and species in the life, including humans, animals, plants, and microbial species (Tang et al., 2019). The sequencing and analyzing for similarity by using BLAST in NCBI compare with the global isolates of this bacterium showed some nucleotide substitutions in some isolates that showed the ability of this bacterium to produce mutations. The results of AST for human and chicken products in this study were agree with (Gharieb et al., 2015) who isolated Salmonella spp. in Egypt from human and poultry meat, that recorded resistance of antimicrobial drugs with Ampicillin 75%, Gentamycin 66.7%, Ciprofloxacin 25 %, and Ceftriaxone 16.7%; while disagree by Tetracycline 100%, Amoxicillin 91.7%, and Trimethoprim 83.3%. The current study was agree with (Diab et al., 2019) who isolated Salmonella spp. in Egypt from human, layer chickens, and chicken eggs, that recorded resistance for Norfloxacin and Ampicillin 68.2%, but disagree by recorded high sensitivity for Gentamicin and Ciprofloxacin 90.9% and 72.7%, respectively. The results of this study with agreement of (Siddique et al., 2021) who isolated S. enterica from poultry and its associated food products from Pakistan, that recorded resistant for Tetracycline 96%, Gentamycin 93%, Trimethoprim 91%, Ampicillin 86%, Ciprofloxacin 19%, and Cefepime 9%; but, disagree by Amoxicillin 81%, Imipenem 12%, and Meropenem 2%. Also, the results agree with (Yang et al., 2020) who isolated this bacterium from retail raw poultry meat in China, that recorded resistance percentage of Ampicillin 55.3%, and 84.1% for each of Ciprofloxacin and Ceftriaxone. This study was disagree with (Rahmani et al., 2011) who isolated this bacterium from birds in Iran, that recorded susceptible to Norfloxacin, Amikacin, and Gentamicin. By observing the results of AST in this study for human and chicken products isolates, we noticed that, there was high resistance percentage (47.37%) for antimicrobial drugs in human isolates compare with the low resistance percentage (15.79%) of antimicrobial drugs in chicken products isolates. Also, showed low susceptibility percentage (52.63%) for antimicrobial drugs in human isolates compare with the high susceptibility percentage (84.21%) for antimicrobial drugs in the chicken products; that is means, the treatment Salmonellosis cases in humans is more difficult and complex compared to the cases of Salmonellosis in the chickens. These variances may be due to a variances in the age, immunity system of human and chickens, over uses of antimicrobial drugs, the improper use of antibiotics in poultry farms, presence of antibiotic residues in foods, non-compliance with the dosage and withdrawal time recommendations nature of nutrition, Salmonella serotypes infection, and hygienic restrictions. Consuming antibiotic residues through animal products may result in the development of antimicrobial drug resistance and lead to increase the pathogenicity and virulence of this pathogen and made the treatments longer and more expensive. Humans, usually, are consuming more antibiotics every time, that is lead to increase the percentage of resistance for antimicrobial drugs (MDR). Resistance genes can be horizontally transferred from Salmonella strains or from other bacterial species to Salmonella bacterium, these additional species may contain antibiotic resistance genes that aren't always present in the Salmonella genetic pool (Hassan et al., 2016). In this study, the results of AST showed that there are varies in the resistance and susceptibility percentages. There are complete resistance percentage (100%) for each of Amikacin, and Gentamicin in both of chicken products, and human isolates. Also, there are complete susceptibility percentage (100%) for each of: Amoxicillin, Pipracillin, Ertapenem, Imipenem, Meropenem, Fosfomycin, and Azithromycin. These results are very important in the selection of the appropriate and optimal drug (Antibacterial agent) for treatment of Salmonella infection in chickens and humans. ConclusionThis study revealed the existence of S. enterica genetic mutations that resulted in differences in the bacteria's molecular traits and resistance to antibiotics. It also revealed similarities between the sequences found in the NCBI that were identified in Iraq and other nations, including the USA, UK, China, Taiwan, Thailand, and Korea. These similarities might result from the importation of foods like chicken products, such as meat and eggs, which might contain S. enterica serovars, or from the entry of people who were sick or carrying these bacteria, particularly foreign workers and military personnel. This could lead to the spread of these bacterial strains from other nations to Iraq and vice versa. It also shows the relationship and route of bacterial infection of S. enterica and the method of transmission of the disease among these bacteria isolated from chicken products and humans. AcknowledgmentThe authors extend their thanks and gratitude to all persons and all staff of all institutions that helped in accomplishing this work. Conflict of interestThe authors declare that there is no conflict of interest. FundingNone. Authors contributionsBoth authors contributed equally in this manuscript. Both authors read and approved the final manuscript. Data availabilityAll data are provided in the manuscript. ReferencesAbdulwahid, M.T., Alobaidi, D.A. and Hamood, M.F. 2017. Evaluation the some biochemical quality and bacterial load of the local and imported chicken meat. Int. J. Sci. Nat. 8(2), 257–260. Abraham, S., O’Dea, M., Trott, D.J., Abraham, R.J., Hughes, D. and Pang, S. 2016. Isolation and plasmid characterization of carbapenemase (IMP-4) producing Salmonella enterica typhimurium from cats. Sci. Rep. 6, 35527. Al-Juburi, L.I. and AL-Sammarraae, I.A. 2023. The protective role of Salmonella typhimurium-whole sonicated killed antigen and Syzygium aromaticum extract on the histopathological changes against its infection in rabbits. Iraqi J. Vet. Med. 46(2), 12–19. Al-Kassie, G.A. 2009. Influence of two plant extracts derived from thyme and cinnamon on broiler performance. Pak. Vet. J. 29(4), 169–173. AL-Khikani, F.H. 2020. Antimicrobial resistance profile among major bacterial pathogens in southern Babil, Iraq. Galician Med. J. 27(3), 36–41. Al-Zuhariy, M.T. 2022. Using T cell lymphokines of hyperimmunized chickens with Salmonella pullorum to protect layer hens against Salmonella pullorum infection. Iraqi J. Vet. Sci. 36(Suppl. I), 223–227. Ansiliero, R., Gelinski, J.M., Samistraro, Q.L., Baratto, C.M., Almeida, C.A. and Locatelli, C. 2021. Pathogenic microbial profile and antibiotic resistance associated with periodontitis. Indian J. Microbiol. 61(1), 55–65. Arsène, M.M., Davares, A.K., Viktorovna, P.I., Andreevna, S.L., Sarra, S., Khelifi, I. and Sergueïevna, M. 2022. The public health issue of antibiotic residues in food and feed: causes, consequences, and potential solutions. Vet. World. 15(3), 662–671. CDC. 2015. National enteric disease surveillance: Salmonella annual report, 2015. Atlanta, GA: Centers for Disease Control and Prevention. Chaudhary, A., Solanki, S., Sharma, D., Singathia, R., Rathore, K., Sain, A. and Gurjar, D. 2022. Isolation, identification, molecular characterization and antibiogram studies of Salmonella spp. isolated from calf diarrhea in and around Udaipur (Rajasthan). Pharma Innov. J. 11(2), 1413–1417. CLSI. 2022. M100 Performance standards for antimicrobial susceptibility testing, 30th ed. CLSI supplement [Electronic]. Malvern, PA: Clinical and Laboratory Standards Institute. Crump, J.A., Sjölund-Karlsson, M., Gordon, M.A. and Parry, C.M. 2015. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 28(4), 901–937. Cuong, N.V., Kiet, B.T., Phu, D.H., Van, N.T., Hien, V.B., Thwaites, G., Mas, J.C. and Choisy, M. 2020. Effects of prophylactic and therapeutic antimicrobial uses in small-scale chicken flocks. Zoonoses Public Health. 68(5), 483–492. Diab, M.S., Zaki, R.S., Ibrahim, N.A. and Abd El Hafez, M.S. 2019. Prevalence of multidrug resistance non-typhoidal salmonellae isolated from layer farms and humans in Egypt. World Vet. J. 9(4), 280–288. Feasey, N.A., Dougan, G., Kingsley, R.A., Heyderman, R.S. and Gordon, M.A. 2012. Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379(9835), 2489–2499. Gharieb, R.M., Tartor, Y.H. and Khedr, M.H. 2015. Non-typhoidal Salmonella in poultry meat and diarrhoeic patients: prevalence, antibiogram, virulotyping, molecular detection and sequencing of class I integrons in multidrug resistant strains. Gut Pathog. 7, 1–11. Hanoun, A.T. and Al-Samrraae, I.A.A. 2019. Isolation and identification of Escherichia coli and Salmonella typhimurium from sheep in Baghdad city. Iraqi J. Vet. Med. 43(1), 124–129. Harb, A., O’Dea, M., Abraham, S. and Habib, I. 2019. Childhood diarrhoea in the Eastern Mediterranean region with special emphasis on non-typhoidal Salmonella at the human–food interface. Pathogens 8(2), 60. Hassan, A.R.H., Salam, H.S. and Abdel-Latef, G.K. 2016. Serological identifi¬cation and antimicrobial resistance of Salmonella isolates from broiler carcasses and human stools in Beni-Suef, Egypt. Beni-Suef Univ. J. Basic Appl. Sci. 5, 202–207. ISO 6579. 2012. Microbiology-general guidance on methods for detection of Salmonella, 4th ed. Geneva, Switzerland: International Organization for Standardization. Jaffer, M.R. 2013. Contamination of local laying hen’s egg shell with Salmonella serotypes. Iraqi J. Vet. Med. 37(1), 13–16. Jajere, S.M. 2019. A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet. World. 12(4), 504. Jammoul, A. and El-Darra, N. 2019. Evaluation of antibiotics residues in chicken meat samples in Lebanon. Antibiotics 8(2), 69–74. Jaslow, S.L., Gibbs, K.D., Fricke, W.F., Wang, L., Pittman, K.J., Mammel, M.K. and Ko, D.C. 2018. Salmonella activation of STAT3 signaling by SarA effector promotes intracellular replication and production of IL-10. Cell Rep. 23(12), 3525–3536. Kanaan, M.H. 2023. Prevalence and antimicrobial resistance of Salmonella enterica serovars enteritidis and typhimurium isolated from retail chicken meat in Wasit markets, Iraq. Vet. World 16(3), 455. Kanaan, M.H.G., Khalil, Z.K., Khashan, H.T. and Ghasemian, A. 2022. Occurrence of virulence factors and carbapenemase genes in Salmonella enterica serovar enteritidis isolated from chicken meat and egg samples in Iraq. BMC Microbiol. 22(1), 1–8. Karasu, K. and Öztürk, E. 2021. Effects of allegations regarding the use of antibiotics and hormones in diets on consumer perceptions, attitudes and behaviors towards broiler meat.consumption. Turk. J. Agricult. Food Sci. Technol. 9(4), 675–682. Kipper, D., Hellfeldt, R.M., De Carli, S., Lehmann, F.K.M., Fonseca, A.S.K., Ikuta, N. and Lunge, V.R. 2019. Salmonella serotype assignment by sequencing analysis of intergenic regions of ribosomal RNA operons. Poult. Sci. 98(11), 5989–5998. Markey, B.K., Leonard, F.C., Achambault, M., Cullinana, A. and Maguire, D. 2014. Clinical veterinary microbiology, 2nd ed. Maryland Heights, MO: Mosby Elsevier, pp: 275–288. Nader, M.I., Rasheed, M.N. and Hammed, H.H. 2015. Molecular identification of Salmonella typhimurium from chicken, meat, and human by PCR. Intl Conf. Med. Genet. Cell. Mole. Biol. Pharm. Food Sci. 2015, 1416–1422. Nakano, M., Yamasaki, E., Ichinose, A., Shimohata, T., Takahashi, A., Akada, J.K. and Kurazono, H. 2012. Salmonella enterotoxin (Stn) regulates membrane composition and integrity. Dis. Models Mechanisms. 5(4), 515–521. Rahmani, M., Peighambari, S.M., Yazdani, A. and Hojjati, P. 2011. Salmonella infection in birds kept in parks and pet shops in Tehran, Iran. Int. J. Vet. Res. 5(3), 145–148. Sahib, A.M., Al-Khalisy, A.F. and Abdulwahid, M.T. 2021. Association of TGF-β2 gene polymorphism with growth rate in local chickens. Iraqi J. Vet. Med. 45(1), 9–16. Siddique, A., Azim, S., Ali, A., Andleeb, S., Ahsan, A., Imran, M. and Rahman, A. 2021. Antimicrobial resistance profiling of biofilm forming non typhoidal Salmonella enterica isolates from poultry and its associated food products from Pakistan. Antibiotics 10(7), 785. Stella, O., Ezenduka, E.V. and Anaelom, N.J. 2020. Screening for Tylosin and other antimicrobial residues in fresh and fermented (nono) cow milk in Delta state, South-South, Nigeria. Vet. World. 13(3), 458–464. Tang, S., Orsi, R.H., Luo, H., Ge, C., Zhang, G., Baker, R.C. and Wiedmann, M. 2019. Assessment and comparison of molecular subtyping and characterization methods for Salmonella. Front. Microbiol. 10, 1591. Thomas, M.E., Klinkenberg, D., Ejeta, G., Van Knapen, F., Bergwerff, A.A., Stegeman, J.A. and Bouma, A. 2009. Quantification of horizontal transmission of Salmonella enterica serovar enteritidis bacteria in pair-housed groups of laying hens. Appl. Environmen. Microbiol. 75(19), 6361–6366. Xu, J., Sangthong, R., McNeil, E., Tang, R. and Chongsuvivatwong, V. 2020. Antibiotic use in chicken farms in northwestern China. Antimicrob. Resist. Infect. Control, 9(10), 72–76. Yang, X., Huang, J., Zhang, Y., Liu, S., Chen, L., Xiao, C. and Wu, Q. 2020. Prevalence, abundance, serovars and antimicrobial resistance of Salmonella isolated from retail raw poultry meat in China. Sci. Total Environ. 713, 136385. Yousif, A.A.R. and Harab, A.A.H. 2011. Isolation and serotyping of Salmonella species in diarrheal children. Univ. Thi-Qar J. Med. 5(1), 149–155. Zubair, A.I., Al-Berfkani, M.I. and Issa, A.R. 2017. Prevalence of Salmonella species from poultry eggs of local stores in Duhok. Int. J. Res. Med. Sci. 5(6), 2468. | ||
| How to Cite this Article |
| Pubmed Style Al-shafee AAJ, Abdulwahid MT. Occurrence, antimicrobial resistance, and molecular characterization of Salmonella enterica from chicken products and human in Wasit Governorate of Iraq. Open Vet. J.. 2024; 14(5): 1117-1129. doi:10.5455/OVJ.2024.v14.i5.5 Web Style Al-shafee AAJ, Abdulwahid MT. Occurrence, antimicrobial resistance, and molecular characterization of Salmonella enterica from chicken products and human in Wasit Governorate of Iraq. https://www.openveterinaryjournal.com/?mno=186671 [Access: January 25, 2026]. doi:10.5455/OVJ.2024.v14.i5.5 AMA (American Medical Association) Style Al-shafee AAJ, Abdulwahid MT. Occurrence, antimicrobial resistance, and molecular characterization of Salmonella enterica from chicken products and human in Wasit Governorate of Iraq. Open Vet. J.. 2024; 14(5): 1117-1129. doi:10.5455/OVJ.2024.v14.i5.5 Vancouver/ICMJE Style Al-shafee AAJ, Abdulwahid MT. Occurrence, antimicrobial resistance, and molecular characterization of Salmonella enterica from chicken products and human in Wasit Governorate of Iraq. Open Vet. J.. (2024), [cited January 25, 2026]; 14(5): 1117-1129. doi:10.5455/OVJ.2024.v14.i5.5 Harvard Style Al-shafee, A. A. J. & Abdulwahid, . M. T. (2024) Occurrence, antimicrobial resistance, and molecular characterization of Salmonella enterica from chicken products and human in Wasit Governorate of Iraq. Open Vet. J., 14 (5), 1117-1129. doi:10.5455/OVJ.2024.v14.i5.5 Turabian Style Al-shafee, Ahmed Abdali Jabor, and Mushtaq Talib Abdulwahid. 2024. Occurrence, antimicrobial resistance, and molecular characterization of Salmonella enterica from chicken products and human in Wasit Governorate of Iraq. Open Veterinary Journal, 14 (5), 1117-1129. doi:10.5455/OVJ.2024.v14.i5.5 Chicago Style Al-shafee, Ahmed Abdali Jabor, and Mushtaq Talib Abdulwahid. "Occurrence, antimicrobial resistance, and molecular characterization of Salmonella enterica from chicken products and human in Wasit Governorate of Iraq." Open Veterinary Journal 14 (2024), 1117-1129. doi:10.5455/OVJ.2024.v14.i5.5 MLA (The Modern Language Association) Style Al-shafee, Ahmed Abdali Jabor, and Mushtaq Talib Abdulwahid. "Occurrence, antimicrobial resistance, and molecular characterization of Salmonella enterica from chicken products and human in Wasit Governorate of Iraq." Open Veterinary Journal 14.5 (2024), 1117-1129. Print. doi:10.5455/OVJ.2024.v14.i5.5 APA (American Psychological Association) Style Al-shafee, A. A. J. & Abdulwahid, . M. T. (2024) Occurrence, antimicrobial resistance, and molecular characterization of Salmonella enterica from chicken products and human in Wasit Governorate of Iraq. Open Veterinary Journal, 14 (5), 1117-1129. doi:10.5455/OVJ.2024.v14.i5.5 |