| Research Article | ||
Open Vet. J.. 2024; 14(8): 1983-1989 Open Veterinary Journal, (2024), Vol. 14(8): 1983–1989 Research Article Monitoring the prevalence of Pseudomonas fluorescens as a spoilage indicator in cow raw milk, teat surfaces, and milk tanksAhmed Hamdi Ahmed1 and Muntaha Ghazi Hassan2*1Nineveh Health Directorate, Ministry of Health and Environment, Mosul, Iraq 2Department of Veterinary Public Health, College of Veterinary Medicine, University of Mosul, Mosul, Iraq *Corresponding Author: Muntaha Ghazi Hassan. Department of Veterinary Public Health, College of Veterinary Medicine, University of Mosul, Mosul, Iraq. Email: fat.ashour [at] uot.edu.ly; mghassan99 [at] uomosul.edu.iq Submitted: 21/05/2024 Accepted: 12/07/2024 Published: 31/08/2024 © 2024 Open Veterinary Journal
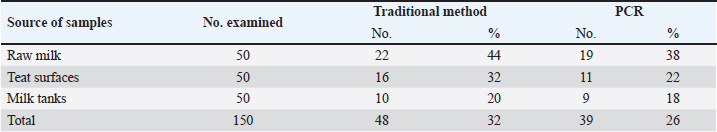
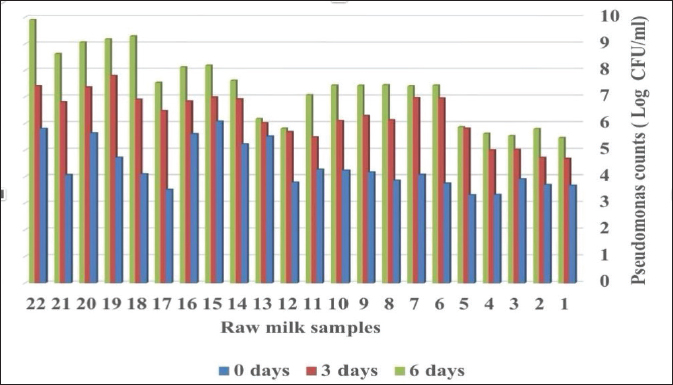
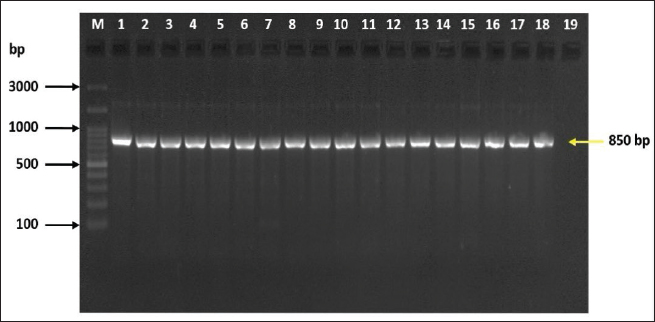
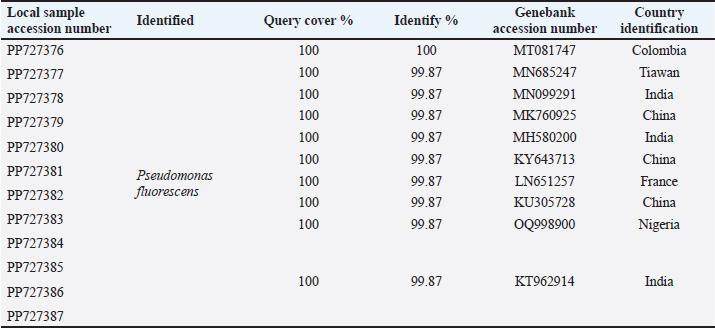
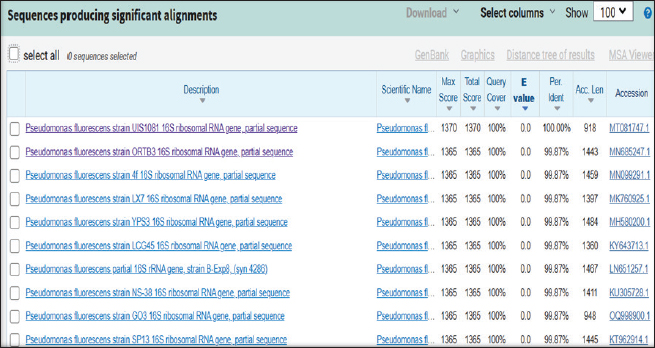
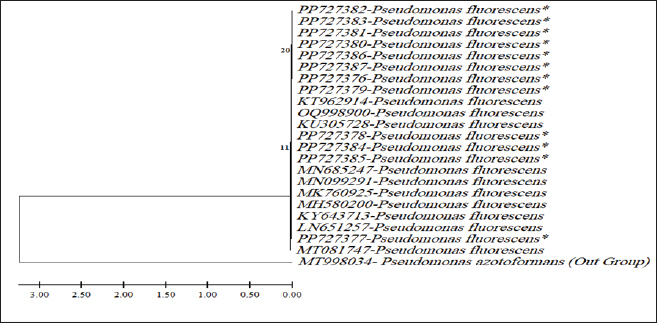
ABSTRACTBackground: Milk and its products are very sensitive to spoilage if they are kept under unsuitable conditions which may provide favorable circumstances for the growth of specific spoilage organisms, Pseudomonas fluorescens accounted as the most dominant indicator for milk spoilage. Aim: This study highlights monitoring the prevalence of P. fluorescens as a spoilage indicator organism in cow raw milk and its contact surfaces represented by teat surfaces and milk tanks in Nineveh province. Methods: A total of 150 samples from cows’ raw milk, teat surfaces, and milk tank swabs were collected from different locations in Nineveh province from October 2023 till February 2024. The Pseudomonas fluorescens were detected by using conventional cultivation methods supported by molecular detection of the target pathogen using the polymerase chain reaction technique. Results: Out of 150 samples, 48 (32%) were positive for the prevalence of P. fluorescens by traditional methods, and 39 (26%) were positive using PCR assay according to the 16SPflu gene yielded a band at 850 bp. The P. fluorescens was recovered at 19 (38%) from raw milk. Teat surfaces revealed a higher isolation rate 11 (22%) compared to milk tanks 9 (18%). The mean counts of Pseudomonas in cows raw milk revealed 4.38, 6.29, and 7.37 log CFU/ml for the 0, 3, and 6 days of storage at chilling temperature. Results of DNA sequencing of the 16SrRNA gene revealed 12 strains recorded in the GenBank nucleotide sequence database. Conclusion: Our results shed light on the risk of P. fluorescens prevalence as a spoilage indicator in raw milk and surrounding surfaces which is inevitable to apply hygienic procedures during milk collecting, processing, and preservation to increase the shelf life of the products and ensure milk safety and consumer health. Keywords: Pseudomonas fluorescens, milk spoilage, teat surfaces, milk tanks. IntroductionMilk has a valuable nutritive value to all ages due to its beneficial nutrients of proteins, carbohydrates, fats, minerals, and vitamins which are necessary for health (Samaržija et al., 2012). Raw milk is very sensitive to contamination with a variety of bacteria from the surrounding environments including the cows, the contaminated milking utensils as well as the soil (Leriche et al., 2004; Bellassi et al., 2021). Usually, raw milk is kept at a low temperature till processing to reduce the multiplication of microbiota, but this is not enough to prevent the proliferation of psychotrophic bacteria groups (Kumar et al., 2019) which can arise aerobically at chilling temperature, especially Pseudomonas fluorescens (Munsch-Alatossava and Alatossava, 2006). Pseudomonas fluorescens belongs to the Pseudomonadaceae family and is considered the most specific spoiling organisms due to their excessive proliferation, therefore it is the most common spoilage indicator in milk and milk products making them unpalatable due to discoloration, off-odor, off-flavor, and slime production (Quigley et al., 2013). All these deterioration signs render the products unfit for human consumption and lead to economic losses in the dairy industry (Santeramo and Lamonaca, 2021). The spoilage activity affects the shelf life of milk due to casein and fat degradation and has been frequently isolated from cheese (Carrascosa et al., 2014; Al-Leboudy, et al. 2015; Stuknytė et al., 2016). Although some Pseudomonas spp. lost their activity after milk pasteurization, but their extracellular enzymes such as proteases remain stable and resist heat treatment leading to degraded milk proteins (Paludetti et al., 2020). Some of P. fluorescens strains possess the ability to form biofilm on equipment’s surfaces which is hard to remove during processing (Ksontini et al., 2013; Rossi et al., 2016). There is obvious spoilage activity when bacterial counts reach to about 107–109 CFU/g of food (Gram et al., 2002). Nowadays milk quality assessment is done through the total bacterial counts before processing which must not exceed 300,000 CFU/ml according to the EC legislation criteria (Regulation EC, 2004). Pseudomonas fluorescens have frequently been used as model organisms to study spoilage mechanisms in dairy products and to evaluate the control approaches that reduce milk contamination (de Oliveira et al., 2015). Therefore, applying hygienic measures through the milking process is necessary to minimize the risk of contamination with psychrotolerant microorganisms (Elmoslemany et al.,2010). To the best of our knowledge, there are few studies investigating spoilage indicators in dairy products in Nineveh province. The current study was highlighted to monitor and detect P. fluorescens from cows’ milk in Nineveh province using the polymerase chain reactions (PCRs) technique. Materials and MethodsSamplesThe research samples included 150 samples as follows, 50 raw milk samples from cows and 50 swabs from each of teat surfaces, and milking tanks. Samples were collected randomly from different regions in Nineveh province between the periods from October 2023 to February 2024. The milk was collected aseptically using sterile bottles, the teat surfaces, and milk tank swabs were taken individually using cotton swabs placed in special sterile tubes containing 5 ml of phosphate buffer solution. All samples were placed in the ice box and transported to the laboratory of the Veterinary Public Health Department, College of Veterinary Medicine, University of Mosul. Isolation and countingRaw milk samples were examined for isolation and counting of psychotropic P. fluorescens at three periods included 0 day then milk samples were cooled at 4oC for 3 and 6 days. Milk samples were diluted 10-fold in 0.1% of sterile peptone water, and 0.1 ml of each dilution was spread on Pseudomonas cetrimide agar (Neogen, USA). Plates were incubated at 25°C for 48 hours, then colonies were counted to calculate as log CFU/ml. The teat surfaces and milk tanks were cultivated on cetrimide agar and plates were incubated aerobically at 25°C for 48 hours. Identifications of bacterial isolatesIdentification of Pseudomonas isolates was done according to biochemical tests represented by the catalase, oxidase, production of pyoverdine, starch hydrolysis, and gelatin liquefaction (Roberts and Greenwood,2008). Vitek2 compact system (Biomerieux, France) were performed to confirm the identification of target bacterium molecular identification of P. fluorescens was used to confirm the diagnosis using PCR technique. DNA extractionPositive colonies were subjected to DNA extraction using a Bacterial DNA preparation kit (Add-bio, Korea) according to the manufacturer’s protocol. Polymerase chain reactionThe presence of P. fluorescens was investigated depending on PCR assay using the 16SPflu gene, a specific primer provided by (Macrogen/Korea) (Table 1). The primer consists of set forward and reverse primers according to (Scarpellini et al.,2004) with a molecular weight of 850 bp. The thermal profile included an initial denaturation of 2 minutes. at 95oC after that, 35 cycles of 94oC for 45seconds, then annealing 56oC for 1 minute. next, extension at 72oC for 1 minute and a final extension of 72oC for 2 minutes. With cooling at 4oC. The products were analyzed by electrophoresis (1.5% agarose gel) provided by (AddBio, Korea) with 3 μl GelRed dye (AddBio, Korea). The pcr products were analyzed in 300mA 75 volts for 1 hour. 4 μl of DNA ladder, 100 bp provided by (GeNet Direx, Korea) was depended as standard. The specific band was identified using the Gel doc EZ image (Bio-Rad, USA). Sequencing of the 16SrRNA geneAfter the PCR products were purified the sequencing of 16SPflu gene was assessed according to Sanger dideoxy sequencing and the Blast algorithm at the NCBI server then phylogenic analysis using ClustalX (NCBI) software programs (Tamura et al.,2021). Statistical analysisThe statistical analysis system was used to detect the differences using SPSS program version 20. The Analysis of Variation was used to assess the significant differences in this study. Ethical approvalNot needed for this study. ResultsThe current study revealed that Out of 150 samples, 39 (26%) were positive for the presence of p. fluorescens strains using PCR techniques compared to 48 (32%) by traditional methods (Table 2). From 50 samples of raw milk19 (38%) were positive for the presence of P. fluorescens, while out of 50 swabs to each of the teat surfaces and milk tanks revealed the recovery rate of P. fluorescens at 11 (22%) and 9 (18%), respectively. With conventional culture and biochemical methods 22 (44%), 16 (32%), and 10 (20%) of isolates were positive for the presence of P. fluorescens in raw milk, teat surfaces, and milk tanks, respectively (Table 3). The counts of Pseudomonas in cows raw milk revealed higher counts of 7.37 ± 0.286 log CFU/ml after 6 days of storage at chilling temperature compared to both of 0 and 3-days storage periods 4.38 ± 0.184 and 6.29 ± 0.192, respectively (Table 4) (Fig. 1). The Pseudomonas counts in milk during storage at chilling temperatures revealed a significant increase at (p < 0.05) compared to the zero day. The PCR results confirmed the detection of P. fluorescens isolates according to the 16SrRNA gene producing product size 850bp (Fig. 2). Sequencing of 16SPflu gene exhibits that strains of P. fluorescens isolated from teat surfaces, raw milk, and milk tank have been submitted to the Genebank database with accession number PP727376. PP727377, PP727378, PP727379, PP727380, PP727381, PP727382, PP727383, PP7273784. PP727385, PP727386, and PP727387, respectively (Table 5) (Fig. 3). Also, Phylogenetic analysis revealed the sequences of our study were similar to those recorded in India, China, Nigeria, Taiwan (Fig. 4). Table 1. Oligonucleotide primer sequence for P. fluorescens used in the current study.
Table 2. Total Prevalence of P. fluorescens in the current study by different assays.
Table 3. Recovery rate of P. fluorescens among different sources using traditional methods and PCR technique.
Table 4. Pseudomonas spp. counts in raw milk stored at 4°C at different periods.
Fig. 1. Total counts of Pseudomonas in raw milk for different storage periods at chilling temperature. DiscussionMilk provides a favorable substrate for bacterial growth, Pseudomonas spp. is one of the important microflora of raw milk and dairy products which may be contaminated via soil and water, they are environmental mastitis-causing pathogens, the importance of Pseudomonas spp. on animal and consumer health related to their many virulence factors as well as to their high resistance to antibiotics (Marchand et al., 2009). Our results showed that the recovery rate of P. fluorescens from raw milk was 38% which accounts high percentage affect milk quality compared to the incidence of P. fluorescens in raw milk from El-Menoufia Governorate 28% (Atia et al., 2022) while it is within the isolation rate(42.66) of P. fluorescens recorded by Meng et al. (2017) in Shaanxi province in China. The percentage of P. fluorescens to the total Pseudomonas spp. was 86.36% compared to the percentage of 75.8% recorded by Du et al. (2022). The isolation rate of P. fluorescens from teat surfaces and milk tank was 22% and 18% respectively, referred that teat surfaces with manual and mechanical milking systems in dairy farms as well as the inner surfaces of milk tank contribute to Pseudomonas spreading (The et al., 2011; Vidal et al., 2017). These results explain the demand for good hygienic practices to control the Pseudomonas spp. contamination in the raw milk stored at chilling temperatures and improve the quality of milk and milk products by controlling the milking process (Leitner et al., 2008; Santana et al., 2020). Also, the results exhibited the Pseudomonas spp. counts ranging from 4.38 to 7.37 log CFU/ml and revealed high levels of Pseudomonas populations in cows’ milk after 6 days of storage at chilling temperatures which similar to some extent to the results obtained by (Lampugnani et al., 2018) in raw milk in Brazil 3–7.1 log CFU/ml while the population of Pseudomonas in raw milk from dairy farms reach to about 4–6 log CFU /ml (Fagundes et al., 2006; Almeida et al., 2017). These results agreed to that both storage time and temperature have a major impact on the growth of Pseudomonas spp. In raw milk (De Jonghe et al., 2011; Lin et al., 2016). The population of Pseudomonas elevated due to their evident activity at low temperatures (Rajmohan et al., 2002; Arslan et al., 2011). Therefore, the contamination of milk with psychotropic bacteria minimizes both milk quality and shelf life of milk and indicates poor hygiene during milking with faulty procedures during storage and transportation of cooled milk (Rajmohan et al., 2002; Ksontini et al., 2013; Al-Rudha et al., 2021). The gene sequencing of P. fluorescens isolates achieved in this study from milk, teat surfaces, and milk tanks were recorded to NCBI indicating a genetic variation and possibility of transmission within the environment (Barton et al., 2013). Thus, monitoring of dairy markets for specific spoilage organisms diminishes the likelihood of Pseudomonas proliferation and prolongs the storage period of the products (Alkhafaje et al., 2022).
Fig. 2. Bands of Electrophoretic profile of the 16SPflu gene of P. fluorescens illustrate Lanes M represent 100 bp DNA marker; lane 1–18, positive samples at 850 bp product size, lane 19 negative control. Table 5. Distribution of Pseudomonas fluorescens based on 16S small subunit ribosomal RNA gene according to BLAST in Genbank of NCBI.
Fig. 3. The identification of the query sample, Pseudomonas fluorescens, in alignment with NCBI gene bank.
Fig. 4. Phylogenic tree of Pseudomonas fluorescens from milk,teat surfaces and milk tanks (*). The P.azotoformans (MT998034-Austria) represent the outgroup. ConclusionDetection of P. fluorescens in cows’ raw milk and milking surfaces indicates the possibility of spoilage arise in milk and may affect consumer health and the high recovery of P. fluorescens requires the application of hygienic conditions during the milking process and handling of milk as well as during milk storage. AcknowledgmentsThe authors would like to thank the University of Mosul, College of Veterinary Medicine in Mosul, Iraq. Conflict of interestThe authors confirm there was no conflict of interest. FundingSelf-funding. Authors’ contributionsThe study was designed, and the manuscript was written and revised by Muntaha Ghazi Hassan and Ahmed Hamdi Ahmed. Both authors approved the final manuscript. Data availabilityAll data supporting the findings of this study are available within the article. ReferencesAlkhafaje, W.K., Olama, Z.A. and Ali, S.M. 2022. Molecular characterization and microbial resistance of different bacterial isolates in some dairy products. Iraq. J. Vet. Sci. 36(2), 333–339. Al-Leboudy, A.A., Nasief A.M., Shaimaa M. and Eltony, S.M. 2015. Occurrence and behavior of Pseudomonas organisms in white soft cheese. Alexand. J. Vet. Sci. 44, 74–79. Almeida, K.M., Bruzaroski, S.R., Zanol, D., Melo, M., Santos, J.S., Aragon Alegro, L.C. and Santana, E.H.W. 2017.Pseudomonas Spp. and P. fluorescens: population in refrigerated raw milk. Ciência Rural, Santa Maria 47(1), 1–5. Al-Rudha, A.M., Khalil, N.K. and Altaai, N.A. 2021.Evaluation of bacterial contaminants and heavy metals in cow and buffalo raw milk sold in Baghdad governorate. Iraq. J. Vet. Sci. 35(Suppl. I-III), 101–105. Arslan, S., Eyi, A. and Özdemir, F. 2011.Spoilage potentials and antimicrobial resistance of Pseudomonas spp. isolated from cheeses. J. Dairy. Sci. 94, 5851–5856. Atia, R.M., Mohamed, H.A., Abo ElRoos, N.A. and Awad, D.B. 2022. Incidence of Pseudomonas species and effect of their virulence factors on milk and milk products. Benha Vet. Med. J. 42, 1–5. Barton, M.D., Petronio, M., Giarrizzo, J.G., Bowling, B.V. and Barton, H.A. 2013.The genome of Pseudomonas fluorescens strain R124 demonstrates phenotypic adaptation to the mineral environ. J. Bacteriol. 195(21), 4793–4803. Bellassi, P., Rocchetti, G., Morelli, L., Senizza, B., Lucini, L. and Cappa, F. 2021.A milk foodomics investigation into the effect of Pseudomonas fluorescens growth under cold chain conditions. Foods 10(6), 1173. Carrascosa, C., Millán, R., Jaber, J.R., Lupiola, P., Rosario-Quintana, C.D., Mauricio, C. and Sanjuán, E. 2014. Blue pigment in fresh cheese produced by Pseudomonas fluorescens. Food Control 54, 95–102. Du, B., Meng, L., Liu, H., Zheng, N., Zhang, Y., Zhao, S., Li, M. and Wang, J. 2022. Diversity and proteolytic activity of Pseudomonas species isolated from raw cow milk samples across China. Sci. Total Environ. 10(Part 3), 156382. Elmoslemany, A.M., Keefe, G.P., Dohoo, I.R., Wichtel, J.J., Strynh, H. and Dingwell, R.T. 2010. The association between bulk tank milk analysis for raw milk quality and on-farm management practices. Prev. Vet. Med. 95, 32–40. Fagundes, C.M., Fischer, V., Silva, W.P., Carbonera, N., Araújo, M.R. and Presença de. 2006. Pseudomonas spp. em função de diferentes etapas da ordenha com distintos manejos higiênicos e no leite refrigerado. Ciência Rural, Santa Maria 36(2), 568–572. Gram, L., Ravn, L., Rasch, M., Bruhn, J.B., Christensen, A.B. and Givskov, M. 2002. Food spoilage--interactions between food spoilage bacteria. Intern. J. Food Microbiol. 78, 79–97. De Jonghe, V., Coorevits, A., Van Hoorde, K., Messens, W., Van Landschoot, A. and De Vos, P. 2011. Influence of storage conditions on the growth of pseudomonas species in refrigerated raw milk. Appl. Environ. Microbiol. 77, 460–470. de Oliveira, G.B., Favarin, L., Luchese, R.H. and McIntosh, D. 2015. Psychrotrophic bacteria in milk: How much do we really know? Braz. J. Microbiol. 46(2), 313–321. Ksontini, H., Kachouri, F. and Hamdi, M. 2013. Dairy biofilm: impact of microbial community on raw milk quality. J. Food Qual. 36(4), 282–290. Kumar, H., Franzetti, L., Kaushal, A. and Kumamoto, D. 2019. Pseudomonas fluorescens: a potential food spoiler and challenges and advances in its detection. Ann. Microbiol. 69, 873–883. Lampugnani, C., Zanatta, M., Montanhini, M., Nero, L. and Bersot, L. 2018. Quantification of psychrotrophic bacteria and molecular identification of Pseudomonas fluorescens in refrigerated raw milk. Food Safe. 86, 1–5. Leitner, G., Silanikove, N., Jacobi, S., Weisblet, L., Bernstein, S. and Merin, U. 2008. The influence of storage on the farm and in dairy silos on milk quality for cheese production. Int. Dairy J. 18, 109–113. Leriche, F., Bordessoules, A., Fayolle, K., Karoui, R., Laval, K., Leblanc, L. and Dufour, E. 2004. Alteration of raw-milk cheese by Pseudomonas spp.: monitoring the sources of contamination using fluorescence spectroscopy and metabolic profiling. J. Microbiol. Methods. 59, 33–41. Lin, H., Shavezipur, M., Yousef, A. and Maleky, F. 2016. Prediction of growth of Pseudomonas fluorescens in milk during storage under fluctuating temperature. J. Dairy Sci. 99(3), 1822–1830. Marchand, S., Vandriesche, G., Coorevits, A, Coudijzer, K., De Jonghe, V., Dewettinck, K., De Vos, P., Devreese, B., Heyndrickx, M. and De Block, J. 2009. Heterogeneity of heat-resistant proteases from milk Pseudomonas species. Int. J. Food Microbiol. 133(1-2), 68–77. Meng, L., Zhang, Y., Liu, H., Zhao, S., Wang, J. and Zheng, N. 2017. Characterization of Pseudomonas spp. and associated proteolytic properties in raw milk stored at low temperatures. Front. Microbiol. 8(8), 2158. Munsch-Alatossava, P. and Alatossava, T. 2006. Phenotypic characterization of raw milk-associated psychrotrophic bacteria. Microbiol. Res. 161, 334–346. Paludetti, L.F., Kelly, A.L. and Gleeson, D. 2020. Effect of thermoresistant protease of Pseudomonas fluorescens on rennet coagulation properties and proteolysis of milk. J. Dairy Sci. 103, 4043–4055. Quigley, L., O’Sullivan, O., Stanton, C., Beresford, T.P., Ross, R., Fitzgerald, G.F. and Cotter, P.D. 2013. The complex microbiota of raw milk. FEMS Microbiol. Rev. 37, 664–698. Rajmohan, S., Dodd, C.E.R. and Waites, W.M. 2002. Enzymes from isolates of Pseudomonas fluorescens involved in food spoilage. J. Appl. Microbiol. 93, 205–213. Regulation (EC). 2004. No 853/2004 of the European Parliament and of the Council of 29 April 2004. laying down specific hygiene rules for on the hygiene of foodstuffs. Available via https://eur-lex.europa.eu/legalcontent/EN/ALL/?uri=celex%3A32004R0853 Roberts, D. and Greenwood, M. 2008. Practical food microbiology. 3rd ed. Oxford, UK: Blackwell Publishing Ltd, pp: 273–274. Rossi, C., Chaves-Lopez, C., Serio, A., Goffredo, E., Goga, B.T.C. and Paparella, A. 2016. Influence of incubation conditions on biofilm formation by Pseudomonas fluorescens isolated from dairy products and diary manufacturing plants. Ital. J. Food Saf. 5(3), 154–157. Samaržija, D., Zamberlin, Š. and Pogačić, T. 2012. Psychrotrophic bacteria and milk and dairy products quality. Mljekarstvo. 62(2), 77–95. Santana, E.H.W., De Luiz, L.L., Pasquim, P.Da.S., Pinto, L.De.F.B., Pereira, F.De.A.B., Gasparini, G.B.F.B., Lorenzetti, E., Bruzaroski, S.R. and Eleodoro, J.I. 2020. Psychrotrophic microorganisms in raw milk and the cheese quality. Res. Soc. Develop. 9(9), 127997217. Santeramo, F.G. and Lamonaca, E. 2021. Food loss–food waste–food security: a new research agenda. Sustainability 13, 4642. Scarpellini, M., Franzetti, L. and Galli, A. 2004. Development of PCR assay to identify Pseudomonas fluorescens and its biotype. FEMS Microbiol. Lett. 236(2), 257–260. Stuknytė, M., Decimo, M., Colzani, M., Silvetti, T., Brasca, M., Cattaneo, S., Aldini, G. and De Noni, I. 2016. Extracellular thermostable proteolytic activity of the milk spoilage bacterium Pseudomonas fluorescens PS19 on bovine caseins. J. Dairy Sci. 99, 4188–4195. Tamura, K., Stecher, G. and Kumar, S. 2021. Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38(7), 3022–3027. The, K.H., Flint, S., Palmer, J., Lindsay, D., Andrewes, P. and Bremer, P. 2011. Thermo-resistant enzyme-producing bacteria isolated from the internal surfaces of raw milk tankers. Int. Dairy J. 21, 742–747. Vidal, A.M.C., Netto, A.S., Vaz, A.C.N., Capodifoglio, E., Gonçalves, A.C.S., Rossi, G.A.M. and Ruiz, V.L.A. 2017. Pseudomonas spp.: contamination sources in bulk tanks of dairy farms. Pesqui Vet Bras 37(9), 941–948. | ||
| How to Cite this Article |
| Pubmed Style Ahmed AH, Hassan MG. Monitoring the prevalence of Pseudomonas fluorescens as a spoilage indicator in cow raw milk, teat surfaces and milk tanks. Open Vet. J.. 2024; 14(8): 1983-1989. doi:10.5455/OVJ.2024.v14.i8.27 Web Style Ahmed AH, Hassan MG. Monitoring the prevalence of Pseudomonas fluorescens as a spoilage indicator in cow raw milk, teat surfaces and milk tanks. https://www.openveterinaryjournal.com/?mno=202568 [Access: January 25, 2026]. doi:10.5455/OVJ.2024.v14.i8.27 AMA (American Medical Association) Style Ahmed AH, Hassan MG. Monitoring the prevalence of Pseudomonas fluorescens as a spoilage indicator in cow raw milk, teat surfaces and milk tanks. Open Vet. J.. 2024; 14(8): 1983-1989. doi:10.5455/OVJ.2024.v14.i8.27 Vancouver/ICMJE Style Ahmed AH, Hassan MG. Monitoring the prevalence of Pseudomonas fluorescens as a spoilage indicator in cow raw milk, teat surfaces and milk tanks. Open Vet. J.. (2024), [cited January 25, 2026]; 14(8): 1983-1989. doi:10.5455/OVJ.2024.v14.i8.27 Harvard Style Ahmed, A. H. & Hassan, . M. G. (2024) Monitoring the prevalence of Pseudomonas fluorescens as a spoilage indicator in cow raw milk, teat surfaces and milk tanks. Open Vet. J., 14 (8), 1983-1989. doi:10.5455/OVJ.2024.v14.i8.27 Turabian Style Ahmed, Ahmed Hamdi, and Muntaha Ghazi Hassan. 2024. Monitoring the prevalence of Pseudomonas fluorescens as a spoilage indicator in cow raw milk, teat surfaces and milk tanks. Open Veterinary Journal, 14 (8), 1983-1989. doi:10.5455/OVJ.2024.v14.i8.27 Chicago Style Ahmed, Ahmed Hamdi, and Muntaha Ghazi Hassan. "Monitoring the prevalence of Pseudomonas fluorescens as a spoilage indicator in cow raw milk, teat surfaces and milk tanks." Open Veterinary Journal 14 (2024), 1983-1989. doi:10.5455/OVJ.2024.v14.i8.27 MLA (The Modern Language Association) Style Ahmed, Ahmed Hamdi, and Muntaha Ghazi Hassan. "Monitoring the prevalence of Pseudomonas fluorescens as a spoilage indicator in cow raw milk, teat surfaces and milk tanks." Open Veterinary Journal 14.8 (2024), 1983-1989. Print. doi:10.5455/OVJ.2024.v14.i8.27 APA (American Psychological Association) Style Ahmed, A. H. & Hassan, . M. G. (2024) Monitoring the prevalence of Pseudomonas fluorescens as a spoilage indicator in cow raw milk, teat surfaces and milk tanks. Open Veterinary Journal, 14 (8), 1983-1989. doi:10.5455/OVJ.2024.v14.i8.27 |