| Review Article | ||
Open Vet. J.. 2022; 12(6): 877-887 Open Veterinary Journal, (2022), Vol. 12(6): 877–887 Review Article Paramyxoviruses in rodents: A reviewFirdaus Mohd-Qawiem1, Abdul Rahman Nawal-Amani1, Farzee Faranieyza-Afiqah1, Abd Rahaman Yasmin2,3, Siti Suri Arshad1, Mohamed Sohaimi Norfitriah2,3 and Saulol Hamid Nur-Fazila1*1Department of Veterinary Pathology and Microbiology, Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Malaysia2Department of Veterinary Laboratory Diagnostics, Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Malaysia 3Laboratory of Vaccines and Immunotherapeutic, Institute of Bioscience, Universiti Putra Malaysia, Serdang, Malaysia Submitted: 25/04/2022 Accepted: 20/10/2022 Published: 20/11/2022 *Corresponding Author: Saulol Hamid Nur-Fazila. Department of Veterinary Pathology and Microbiology, Faculty of Veterinary Medicine, Universiti Putra Malaysia, Serdang, Malaysia. Email: nurfazila [at] upm.edu.my © 2022 Open Veterinary Journal
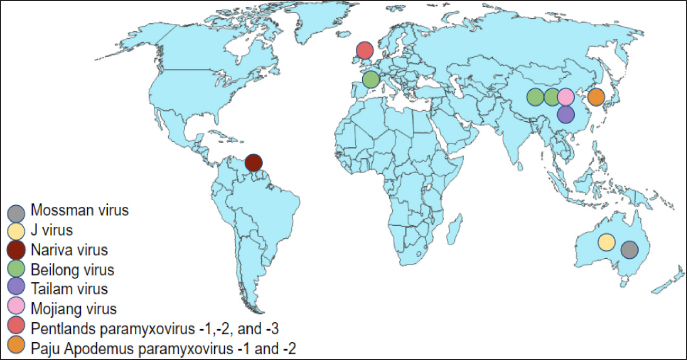
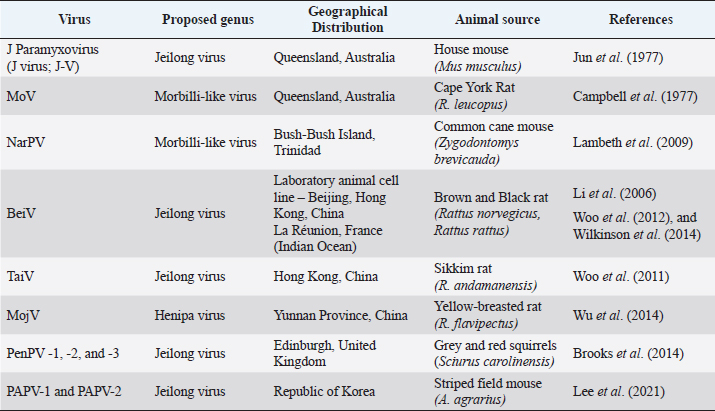
AbstractParamyxoviruses have been shown to infect a wide range of hosts, including rodents, and humans. Several novel murine paramyxoviruses have been discovered in the last several decades. Although these viruses are unclassified, they are recognized as Beilong virus, Mojiang virus (MojV), and Tailam virus in rats, Jeilongvirus, Nariva, Paju Apodemus paramyxovirus-1 and -2 in mice, and Pentlands paramyxovirus-1, -2, and -3 in squirrels. These paramyxoviruses were reported mainly in China and a few other countries like Australia, the Republic of Korea, Trinidad, and France. In June 2012, it becomes a great concern in China whereby, three miners were reported dead potentially caused by a novel zoonotic MojV, a henipa-like virus isolated from tissue samples of rats from the same cave. Rats are considered to be natural hosts for the MojV from the literature research. The classified paramyxovirus, Sendai virus in rodents is also reviewed. Paramyxoviruses infection in rodents leads to respiratory distress such as necrotizing rhinitis, tracheitis, bronchiolitis, and interstitial pneumonia. Infections caused by paramyxoviruses often spread between species, manifesting disease in spillover hosts, including humans. This review focuses on the paramyxoviruses in rodents, including the epidemiological distributions, transmission and pathogenesis, clinical manifestations, diagnostic methods, and control and prevention of paramyxoviruses infection to provide a better understanding of these highly mutating viruses. Keywords: Beilong virus, Jeilongvirus, Mojiang virus, Paramyxoviruses, Sendai virus. IntroductionThe family Paramyxoviridae is subdivided into four subfamilies: Avulavirinae, Rubulavirinae, Metaparamyxovirinae, and Orthoparamyxovirinae. To date, the subfamily Orthoparamyxovirinae is comprised of eight genera: Respirovirus, Aquaparamyxovirus, Ferlavirus, Henipavirus, Narmovirus, Jeilongvirus (J-V), Salemvirus, and Morbillivirus (Rima et al., 2019). These deadly family viruses are known to infect a wide range of host species such as cattle, sheep, horses, bats, rodents, and humans, leaving a massive impact on both humans and animals, in perspectives of public health and economic loss (Normile, 2008; Bellini and Rota, 2011). The entry of as-yet-unknown paramyxoviruses from wildlife reservoirs into human populations has been highlighted in zoonotic outbreaks of the very fatal Hendra virus and Nipah virus (Thibault et al., 2017). Bats and rodents have a wide range of species and are accustomed to congregating in big groups are significant drivers of cross-species viral transmission (Wang et al., 2008; Brook and Dobson, 2015; Han et al., 2015). According to Drexler et al. (2012), the shifts of paramyxovirus host are most prevalent in other mammalian species from bats. New paramyxoviruses possibly continue to develop mostly from bat reservoir spillover. Furthermore, among RNA viruses, paramyxoviruses become the greatest degrees of cross-species transmission (Zeltina et al., 2016). The family continues to emerge after several novel paramyxoviruses like the J-V, Mossman virus (MoV), and Nariva virus (NarPV) that discovers over the last few decades (MacLachlan et al., 2011). Many paramyxovirus diseases spread between species, causing re-emergences of illnesses in spillover hosts such as humans. Some viruses potentially cross species barriers and establish infection, raising public health concerns (Natasha et al., 2022). Rodents are considered to have given rise to several paramyxoviruses including J-V, MoV, NarPV, Beilong virus (BeiV), Tailam virus (TaiV), Mojiang virus (MojV), Pentlands Paramyxovirus-1, -2, -3 (PenPV-1,-2,-3), Paju Apodemus Paramyxovirus-1, -2 (PAPV-1, -2), and Sendai virus (SeV). Although novel variants are being identified, the interaction between the virus and host is not extensively described. This review article focuses on the etiological agent, virus replication, epidemiology, unclassified and classified paramyxoviruses, including the transmission and pathogenesis, clinical manifestations, diagnostic methods, and control and prevention of paramyxoviruses infection in rodents to provide insight into these highly mutating viruses. Paramyxoviruses PropertiesParamyxoviruses are single-stranded RNA viruses with negative-sense RNA genomes that are enveloped. The virions have a diameter of 150 nm or higher (up to 500 nm), pleomorphic, but in vitreous ice, they are often spherical. The viruses are composed of a lipid envelope encasing a nucleocapsid. The envelope is composed of two transmembrane glycoproteins that are formed directly from the plasma membrane of the host cell through the process of budding. Currently, there are no known vectors for the virus, which allow it to spread horizontally through direct contact and airborne transmission (Rima et al., 2019). Virus ReplicationParamyxoviruses reproduce in the cytoplasm of infected cells and continue to replicate in the presence of actinomycin D and enucleated cells, demonstrating that nucleus functionalities are not required. The corresponding attachment proteins (HN, H, and G) identify ligands on the target cell’s surface and are compatible with them. Following attachment, the mature F protein facilitates the plasma membrane fused with the viral envelope. The ribonucleoprotein (RNP) is subsequently released into the cytoplasm, where the N, P, and L proteins commence transcription of mRNAs encoding the viral proteins from the genomic viral RNA, allowing mRNA synthesis to begin before the synthesis of de novo protein. The P and L proteins of the polymerase complex begin the synthesis of RNA at the genomic RNA of 3’ end and transcribe the genome sequentially into 6 to 10 distinct capped and polyadenylated mRNAs. This termination-reinitiation mechanism regulates mRNA synthesis in such a way that the amount of individual mRNAs reduces as the genome moves further from the 3’ end. Transcription of a promoter region at the 3’ end of the genomic RNA begins when the N protein concentration reaches a threshold level, and then it attaches to the nascent RNA chain. The polymerase disregards message termination signals, resulting in the whole positive-sense antigenome synthesis. The N protein-encapsulated antigenome is used to generate negative-sense genomic RNA. The mRNA synthesis in the second phase is subsequently initiated from the freshly synthesized genomic RNA, significantly increasing the viral protein production (MacLachlan and Dubovi, 2017). Syncytia formation is a common feature of many paramyxovirus infections in nonpolarized cell cultures, but it is uncommonly seen in polarized cell culture systems. It is a distinguishing feature in paramyxovirus infections of some animals, but not all. According to MacLachlan and Dubovi (2017), acidophilic cytoplasmic inclusions consisting of RNP structures are the hallmark of paramyxovirus infections, and while morbilliviruses replicate exclusively in the cytoplasm, they also create acidophilic intranuclear inclusions composed of nuclear elements and N protein complexes. Hemadsorption is a characteristic of paramyxoviruses that include the HN protein, as well as some morbilliviruses. EpidemiologyNovel paramyxoviruses are continuously being identified in rodent species. Rodents carry at least eight paramyxoviruses such as J-V, MoV, NarPV, BeiV, TaiV, MojV, PenPV-1,-2,-3, and PAPV-1, -2 and classified one of SeV (Sasaki et al., 2014). The paramyxoviruses have been discovered since early 1977 until recently in 2021 in different rodent species over the world, including Trinidad, the United Kingdom, France, Korea, Australia, and China (Fig. 1). The epidemiological distribution of emerging paramyxoviruses in rodents is summarized in Table 1. Unclassified ParamyxovirusesJeilongvirusThe J-V was discovered in 1977, in a kidney autoculture of a dead house mouse, Mus musculus in Northern Queensland, Australia (Jun et al., 1977). Severe hemorrhagic pneumonia was observed in the infected mice. The J-V was used to infect human MRC5, Hep2 cell lines, pig kidney (PS), baby hamster kidney (B11K21), and monkey kidney (Vero) cells cytopathic effects (CPE). The CPE were characterized by the loss of a single monolayer, the formation of the vacuolated syncytium, and extensive eosinophilic cytoplasmic inclusions. CPE was not detected in human liver cells from the Hela or Chang lines, bovine kidney cells (MDBK), or rat fibroblasts. The paramyxovirus virion morphology demonstrated by electron microscopy (EM) revealed a 15-nm-diameter herringbone-shaped nucleocapsid structure. J-V haemagglutinin was not detectable in mice, rats, guinea pigs, rabbits, chickens, geese, pigs, sheep, cows, horses, or human Type O erythrocytes at various temperatures with complement fixation antigen. The genomic analysis of J-V was also documented previously (Jack et al., 2005). Mossman virusThe MoV was discovered in two species of rats: Rattus leucopus in Mossman, Australia, in 1970, and Rattus fuscipes in Mossman, Australia, in 1971 (Campbell et al., 1977). MoV was recognized as a novel paramyxovirus based on its CPE in cell culture, the virions and nucleocapsids morphology on EM, and reduced reactivity with antisera to a wide range of viruses, including NarPV and J-V. The natural transmission mechanism of MoV is unknown. Antibodies to MoV were found in rats of a survey study in Queensland from 1998 until 1999, indicating that MoV or a closely related virus was still circulating in the rodent populations around 30 years after its discovery in the same area. This virus was the first of a unique group of mouse paramyxoviruses to undergo molecular sequencing. According to genomic and protein analysis, MoV is previously known as a novel paramyxovirus (Miller et al., 2003).
Fig. 1. Epidemiological distribution of unclassified paramyxoviruses. Table 1. Epidemiological distribution of emerging paramyxoviruses in rodents.
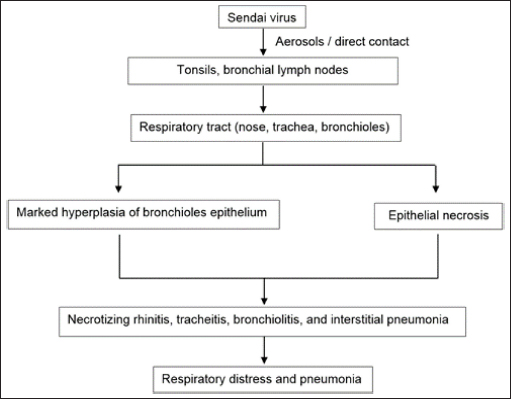
Nariva virusThe NarPV was discovered in the Trinidadian mouse, Zygodontomys b. Brevicauda, in the early 1960s (Karabatsos et al., 1969; Tikasingh et al., 1996). In Vero and BHK cells, the virus replicated in the suckling mouse brain and generated syncytia. It was identified as a paramyxovirus mostly due to the structure of its nucleocapsids, which had a diameter of 20 nm and a mean length of 1.8 nm. The virion is a pleomorphic sphere with enveloped surface projections. In contrast to morbilliviruses, which produce nuclear and cytoplasmic inclusion bodies in virus-infected cells, NarPV creates only cytoplasmic inclusion bodies in the infected cells (Walder, 1971). The genome of the NarPV is 15,276 nucleotides (nt) long, follows the rule of six in paramyxovirus, and is organized similarly to that of the majority of paramyxoviruses (Lambeth et al., 2009). Suckling hamsters infected with the NarPV developed acute necrotizing encephalitis, as well as significant levels of infectious virus and virus antigen in the brain. However, weanling hamsters had only trace levels of infectious virus at the early stage of the disease, when they were healthy. Then, they developed a non-productive infection with persistent indications of viral antigen but no detectable infectious virus when they became clinically symptomatic. Weanlings died earlier than sucklings due to cellular necrosis at the cerebral parenchyma. Beilong virusBeiV was first discovered in a kidney mesangial cell line of humans, although the origin of the virus was presumed to be a kidney mesangial cell line of rats (Li et al., 2006). BeiV and J-V, together with MoV and Tupaia Paramyxovirus, have always been classified as belonging to the morbillivirus and respirovirus genera in a phylogenetic tree (Li et al., 2006). BeiV’s entire genome sequence was discovered to be the largest, measuring 19,212 nt and being 258 nt longer than the next largest member of the Paramyxoviridae, J-V (18,954 nt). Woo et al. (2012) found that BeiV could be amplified in a rat kidney mesangial cell but not in a human kidney mesangial cell. The strains of BeiV had already been discovered in rats from China. Asymptomatic brown rats (Rattus norvegicus) and black rats (Rattus rattus) were used to extract kidney, spleen, and respiratory and anal swabs. BeiV was found in samples of kidney and spleen but undetected in respiratory or anal swabs, suggesting it causes systemic infection but is eliminated through urine. BeiV and its variants are abundant in brown and black rats, but the mode of transmission is yet unknown (Woo et al., 2012). The previously mentioned discoveries suggest that rodent is the natural host of BeiV (Sasaki et al., 2014). It was found at high concentrations in numerous organs, with the most abundantly found in the kidney, followed by the spleen, liver, lung, and heart, although the exact origin of the viral host is currently unknown (Chen et al., 2020). Similarly, the researchers hypothesized that BeiV might cause systemic infection in a host. It could be transmitted to rodents by biting or blood-sucking vectors, despite the lack of direct evidence of BeiV detection in blood samples. Tailam virusA novel paramyxovirus of TaiV was identified in Tai Lam, Hong Kong, in the kidneys and spleens of Sikkim rats (Rattus andamanensis) between 2008 and 2009 as part of epidemiological investigations on paramyxoviruses in rodents. The virus was revealed to be related to, but distinct from, the BeV and J-V viruses (Woo et al., 2012). It comprises an eight-gene virus with a genomic size of 19,152 nt (Woo et al., 2011). In China, the TaiV was found in R. andamanensis with a complete genome sequence documented (Woo et al., 2011). Mojiang virusAn unknown source was responsible for the deaths of three persons who were working at an abandoned mine in Mojiang Hani Autonomous County in Yunnan Province, China, in 2012 (Wu et al., 2014). The occurrence of new zoonotic illnesses in natural hosts in the cave was explored by the researchers half a year later. Anal swab samples collected from bats (Rhinolophus ferrumequinum), rats (Rattus flavipectus), and musk shrews (Crocidura dracula) from the mine were utilized for metagenomic sequencing of viral nucleic acids. The researchers detected 38 sequence reads that were categorized as Henipavirus based on the non-redundant protein alignments that share only a few nt and amino acid sequences with known henipaviruses (Wu et al., 2014). MojV is a henipavirus that has a genome length of 18,404 nt, which is similar to the length of henipavirus genomes. The relationship between MojV and other paramyxoviruses was established by the development of phylogenetic trees based on the N and L protein sequences, respectively. It was determined that MojV was distinct from the other clusters and that it belonged to the Henipavirus genus along with the other four members. Based on the similarities between the genomes of MojV and other henipaviruses, they believed that MojV should be classified as a unique species closely related to the Henipavirus species. Three of the nine anal swab samples taken from R. flavipectus rats tested positive for MojV, as a tissue sample taken from one of the three MojV-positive R. flavipectus rats tested positive. Three MojV-positive anal swab samples cultured in Vero E6, Hep2, and BHK21 cells for virus isolation showed no cytotoxic effects and no viral multiplication. In the investigation, the discovery of MojV, a henipa-like virus with a rodent origin, was verified to take place in China. Rats of the genus R. flavipectus are the natural reservoir for the MojV virus. Due to this discovery, henipaviruses are known to infect a wider range of mammalian hosts than previously assumed, and they are not restricted to bats alone. Therefore, the MojV isolated from rats, R. flavipectus, possibly caused the deaths of three miners in China (Wu et al., 2014). Pentlands paramyxovirus-1, -2, and -3Three novel rodent paramyxoviruses, PenPV -1, -2, and -3, have been discovered in grey and red squirrels in the United Kingdom. PenPV is belonging to the subfamily Paramyxovirinae subfamily, although it did not partition with unclassified rodent paramyxoviruses such as J-V, BeiV, or TaiV (Brooks et al., 2014). PenPV-1 was detected in the kidney cells of a grey squirrel, Sciurus carolinensis, which died in a car collision in Edinburgh, United Kingdom, in 2014. It is closely related to two additional paramyxoviruses detected in red squirrels, PenPV-2 and PenPV-3, according to the characteristics of cell culture growth, the virion and nucleocapsid ultrastructure under electron microscopy, and the 1,788 nt analysis of the virus genome. PenPV-1 can infect red or grey squirrels infected with PenPV-2 or -3, but it has been unproven. PenPV-1 infection of grey squirrel’s kidney cells in cell culture resulted in the development of syncytia, but they did not seem to be a barrier to their lives, because they lived for 13 weeks without signs of virus-induced apoptosis. When PenPV-1 infected BHK-21 cells, however, the majority of the cultured cells died after 48 hours. It indicates that PenPV-1 infection has a variety of effects, depending on the infected cell type (Brooks et al., 2014). According to Brooks et al. (2014), the fact that viruses in one of the red squirrels are closely associated with paramyxoviruses obtained in free-ranging bats may indicate transmission of the cross-species episode, although the exact mode of transmission is unknown. Once again, it’s unclear if their respective hosts could be infected by these viruses. Paju Apodemus paramyxovirus-1 and -2PAPV-1 and PAPV-2 were discovered in the Republic of Korea’s Apodemus agrarius (the striped field mouse) in June 2021 (Lee et al., 2021). It was found in 108 (or 13.1%) of the A. agrarius individuals tested positive, accounting for 10.6% and 2.6% for PAPV-1 and PAPV-2, respectively (Lee et al., 2021). When the complete genomes of the PAPVs were sequenced, it was discovered that they are genetically unique in the Paramyxoviridae family, based on phylogenetic analysis. Rodent paramyxoviruses of J-V, BeiV, and TaiV had the same genomic organization as these viruses. The PAPV-1 genome (19,716 nt) shares the same genomic properties and length as the Pohorje Myodes paramyxovirus 1 (PMPV-1) that was found in the kidney of a bank vole (Myodes glareolus) (Vanmechelen et al., 2018), which is the biggest group of rodent-borne paramyxoviruses that have been found to date. The TM and SH genes, together with the high G gene size, are all factors contributing to this phenomenon. PAPV-2’s total genome was 17,475 nt in length, about 2 kb less than PAPV-1, as a result of the SH gene and short G gene deletion. According to their distinct traits and evolutionary distance from other paramyxoviruses, it has been suggested that these viruses form a new genus in the Paramyxoviridae family (Lee et al., 2021). Classified ParamyxovirusesSendai virusTransmission and pathogenesisA member of the Respirovirus genus, the SeV (murine respirovirus) is thought to be the causative agent of lung inflammation in rat species over the world (Faísca and Desmecht, 2007). It causes severe respiratory illness in adult mice (Appell et al., 1971) and, to a lesser degree, rats, and other experimental animals (MacLachlan et al., 2011). Depending on a variety of factors including age, strain susceptibility, husbandry, transportation, and co-pathogens, infection with the SeV can manifest itself in different ways (Nicklas et al., 2012). Fischer rats infected with the SeV showed much greater viral titers and significantly longer viral replication than Brown Norway rats (Sorden and Castleman, 1991). SeV pneumonia is very infectious in the DBA mouse strains, while C57BL/6/J, SJL/J mice, and some outbred stocks become relatively resistant (Parker et al., 1978; Nicklas et al., 2012). This results in mild to moderate symptoms in older and genetically resistant strains of mice, since the virus is unable to reach the distal airways before the immune response is activated, resulting in less severe symptoms in young animals (MacLachlan et al., 2011). Generally, paramyxoviruses enter the host through the respiratory tract, where they replicate and spread throughout the body. However, certain paramyxoviruses such as Newcastle disease and morbilliviruses can infect the gastrointestinal tract (Samal, 2008). SeV replicates in the cytoplasm of the host cell to a significant extent. A disulfide bond connects the two fusion (F) protein subunits (F1 and F2) after the F0 protein has been processed by the serine protease before one of the three functional proteins of the virion envelope is activated. Cleavage is induced by certain proteases that exist naturally in the host, primarily in the respiratory system. “Tryptase Clara” is a serine protease that is hypothesized to have a role in the virus’s ability to infect the espiratory tract (Parker and Ritcher, 1982). The protease may be found along the terminal bronchioles and can cleave the protein F, allowing the virus to become active and spread (Kido et al., 1992; Tashiro et al., 1992). SeV is extremely contagious and can be transmitted by aerosols (Nicklas et al., 2012) or direct contact (Barthold et al., 2016) between infected individuals. Rapid transmission frequently occurs among rat populations before the introduction of individually ventilated cages (Nicklas et al., 2012). During infection, type II pneumocytes and respiratory epithelial cells in the nose, trachea, and bronchioles are targeted by the virus; however, infected cells are not always destroyed (MacLachlan et al., 2011). The pathogenesis of paramyxovirus infection in rodents is portrayed in Figure 2.
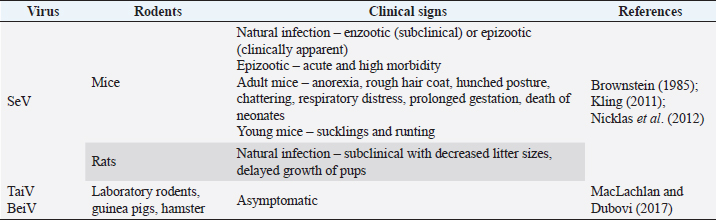
Fig. 2. Pathogenesis of SeV infection in rodents. Clinical manifestationsThe SeV also known as murine parainfluenza virus-1 has been associated with respiratory sickness in laboratory mice, rats, hamsters, and in guinea pigs (Barthold et al., 2016). Table 2 summarizes the clinical signs of paramyxovirus infection in rodents. Animals like other rodents (guinea pigs, beavers, squirrels, etc.), and lagomorphs (rabbits, etc.) are frequently infected with viruses that are mild or asymptomatic (MacLachlan et al., 2011). Cytotoxic T lymphocytes assault virus-infected cells during infection at the "immune" phase, resulting in lesions in respiratory tracts including necrotizing rhinitis, tracheitis, bronchiolitis, and interstitial pneumonia, among other manifestations. Clinical indications manifest themselves in completely immunocompetent animals, with various degrees of morbidity and death according to the age, strain, and immunocompetence of the animals involved. Instead of pathognomonic immune-mediated necrotizing bronchiolitis, nude mice develop long-term progressive interstitial pneumonia, a type of chronic lung infection (Ward et al., 1976). Furthermore, rats infected by SeV produce fewer litter sizes and poor pup development (Kling, 2011). Enzootic infection in rats can manifest itself in two distinct ways: as enzootic (subclinical) infection or as epizootic (clinical) as discussed by Nicklas et al. (2012). The development of latent infection follows an enzootic pattern. Mice in breeding colonies obtain infection promptly after weaning due to low levels of antibodies in the maternal serum after birth. The pattern of the epidemic, on the other hand, is often acute, contributing to increased morbidity and mortality, with almost all susceptible animals getting infected within a short period of time. Between 2 and 7 months after the virus is initially introduced to a colony, sick animals might die or become sub-clinically infected. Diagnostic Methods of Paramyxoviruses InfectionSerological diagnosisSeV infection in rats is best diagnosed by Indirect Enzyme-Linked Immunosorbent Assay (ELISA) or Multiplexed Fluorometric ImmunoAssay due to their sensitivity to detect the early and low antibody titers (Kling, 2011). Rothenburger et al. (2015) used an ELISA with minimal alterations to detect respiratory infections in rats. Serum samples from 139 rats were tested for the bovine respiratory syncytial virus, a pneumovirus related to murine pneumonia virus, and the bovine parainfluenza virus 3, a virus related to the SeV. The results revealed that lysed Vero cells infected with these isolates generated partially purified antigens, with lysed uninfected Vero cells acting as a positive control. No evidences for the rat coronavirus, SeV or murine pneumonia virus were found. However, the negative finding is possibly due to the small sample size employed in the testing. In laboratory rodent colony settling, ELISA and immunofluorescence techniques are most commonly used (MacLachlan and Dubovi, 2017). Antibodies can be detected 7 days after infection, and their presence is often linked to the beginning of immune-mediated necrotizing bronchiolitis and pneumonia. The presence of the virus is further confirmed by the immunofluorescence or immunohistochemical staining of infected monolayers in the embryonated eggs or a variety of cell culture methods including monkey kidney, Vero, and BHK-21 cells with trypsin in the culture conditions. Table 2. Clinical signs of paramyxoviruses infection in rodents.
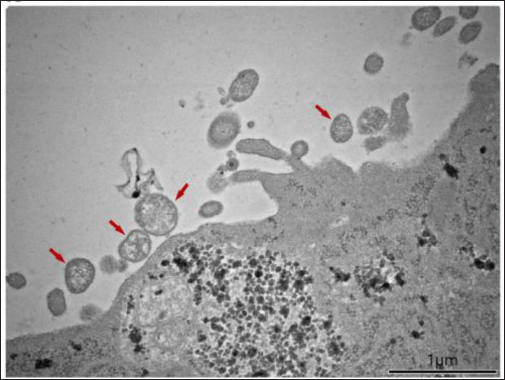
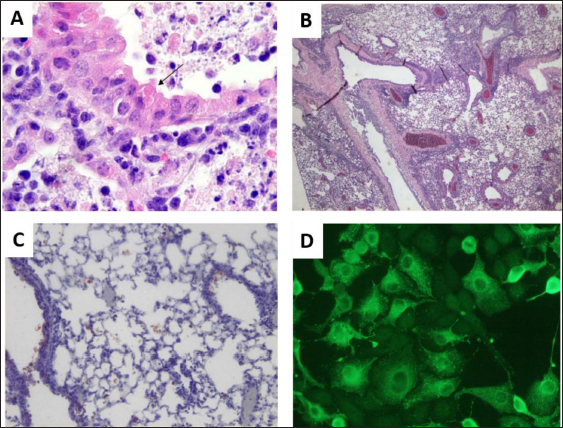
Reverse-transcriptase polymerase chain reaction (RT-PCR)The method of RT-PCR is currently the gold standard for rapid testing and confirmation of the isolates (MacLachlan and Dubovi, 2017). Chen et al. (2020) reported the spread of BeiV across the common species of wild rodents and shrews in China. The virus was screened using modified primers, and a 440-bp fragment of the large (L) gene was amplified by PCR, followed by reverse transcription-PCR. A total of 7,168 samples were collected and evaluated from 2005 wild animals (rodents, shrews, and Mustelidae). BeiV was identified in large quantities in the kidney, spleen, liver, lung, and heart. But, it was not found in any samples retrieved from the gut. Previous studies revealed that BeiV was possibly detected in kidney and spleen samples, but not in respiratory or anal swabs (Woo et al., 2016). Similarly, paramyxovirus viral RNA was not found in any fecal samples of bats and rats in the Mekong Delta region of Southern Vietnam (Berto et al., 2017). This could be owing to the study’s use of fecal samples, which were acquired incorrectly. Chen et al. (2020) determined that BeiV is a naturally occurring and extensively diffused virus among rats in China, based on the positive results in three out of four rodent groups (Cricetidae, Muridae, and Sciuridae). Virus isolation and electron microscopyIsolation of novel PAPV-1 was carried out by Vero E6 cells using rodent kidney tissues, according to Lee et al. (2021). CPE were monitored under inverted microscopy on a regular basis. Following that, samples were obtained 1, 3, 5, and 7 days after infection. Herein, paramyxovirus isolates of infected animals revealed a cytotoxic effect of syncytia formation in Vero E6 cells which was not shown in this article. For confirmation, the first isolate of PAPV-1 was passaged twice for 14 days after inoculation. A transmission electron microscope was then used to examine PAPV-1 (Fig. 3). Microscopic lesionsThe most common lesions induced by SeV infection are tracheitis and bronchopneumonia although the degrees of tissue reaction are varying but the lesions develop quickly and recover swiftly (Faísca and Desmecht, 2007). Throughout the course of time, lesions spread throughout the respiratory system. Epithelial cells exhibit tiny eosinophilic cytoplasmic aggregates with a clear halo, measuring 1 to 3 microns in diameter (Fig. 4A). It appears as proliferative bronchiolitis in the lower airways, with substantial epithelial hyperplasia, cilia loss, giant cell development, and desquamation in the lower airways and upper airways (Itoh et al., 1991). Alveoli are heavily filled with inflammatory cells, proteinaceous material, and cellular debris as they extend from the altered terminal bronchioles of the lungs (Fig. 4B). SeV infection is often seen near an airway (Brownstein et al., 1981), but in severe situations, inflammation infiltrates more peripheral locations and even whole lobes (Percy et al., 1994). However, it cannot depend solely on gross and histological evidence alone because the lesions are frequently minor and non-pathognomonic. To improve the specificity and sensitivity of histopathology, other methods such as immunohistochemistry (Fig. 4C) and immunofluorescence (Fig. 4D) can be utilized to identify SeV-specific antigens. Control and PreventionParamyxoviruses in particular are known to cause infections in humans. According to MacLachlan et al. (2011), depopulation, cleaning of the facilities, and screening of incoming animals are necessary as preventive measures after the disease are detected. Immune-competent animals that recover from infection may not carry the virus, but antibodies can be identified in their blood and persist in their bodies for the rest of their lives. A variety of techniques including foster nursing, embryo transfer, and segregating immunocompetent breeding mice would give birth to uninfected pups but temporarily seropositive might be utilized to assist afflicted colonies. When it comes to immunodeficient mice, cesarean or embryo transfer derivation is preferable since the virus is only capable of infecting the respiratory system. Every offspring must be thoroughly screened before breeding or reintroducing animals into uninfected groups in order to assure effective re-derivation of the pathogens.
Fig. 3. PAPV-1 particles using a transmission electron microscope.
Fig. 4. SeV infection of a mouse lung. (A) Small eosinophilic cytoplasmic bodies (arrow) are seen in an airway epithelial cell (H & E, 200×). (B) Proliferative bronchitis is characterized by significant hyperplasia of epithelial cells, cilia loss, and desquamation of the bronchial epithelium (H & E, 40×). (C) Positive cells for the SeV antigen in the alveolar spaces and bronchial epithelium stained with immunoperoxidase, 100×. (D) Positive LLC-MK2 cells for the SeV antigen via immunofluorescence using a rabbit polyclonal anti-Sendai antibody, 400×. ConclusionIn conclusion, paramyxovirus infections in rodents are highly contagious and potentially harbor zoonotic potential, raising public health concerns. The shedding of infectious virus at the first 2 weeks of infection seems to be transmitted through direct contact, respiratory aerosol, and contaminated fomites. The clinical signs in paramyxovirus-infected animals are dyspnea, chattering, prolonged gestation, reduced growth, and even young mice death. Screening of incoming rodents, isolation, and depopulation of infected rodents should be performed as control and preventive measures for paramyxovirus infection. As a recommendation, thorough research should be conducted to gain more information on the virus and host interactions in terms of transmission, pathophysiology, and the natural host of the paramyxoviruses due to reported zoonotic cases. Although the knowledge of the virus and host interaction is scarce, further study on the zoonotic potential of paramyxovirus infections due to the possibility of the virus continuously mutating among the rodent species is needed. AcknowledgmentsThis work was supported by UPM as a research grant provider (UPM/GP-IPS/2021/9705100). Conflict of interestThe authors declare that there is no conflict of interest. Authors’ contributionsNAAR reviewed the previous papers. NFSH wrote the article and edited the final draft of the manuscript. All authors read and approved the final manuscript. ReferencesAppell, L., Kovatch, R., Reddecliff, J.M. and Gerone, P.J. 1971. Pathogenesis of Sendai virus infection in mice. Am. J. Vet. Res. 32(11), 1835–1841. Barthold, S.W., Griffey, S.M. and Percy, D.H. 2016. Pathology of laboratory rodents and rabbits, 4th ed. Hoboken, NJ: Wiley Online Library. Bellini, W.J. and Rota, P.A. 2011. Biological feasibility of measles eradication. Virus. Res. 162(1–2), 72–79. Berto, A., Anh, P.H., Carrique-Mas, J.J., Simmonds, P., Van Cuong, N., Tue, N.T., Van Dung, N., Woolhouse, M.E., Smith, I., Marsh, G.A., Bryant, J.E., Thwaites, G.E., Baker, S., Rabaa, M.A. and Consortium VIZIONS. 2017. Detection of potentially novel paramyxovirus and coronavirus viral RNA in bats and rats in the Mekong Delta region of southern Vietnam. Zoonoses Public Health 65(1), 30–42. Brook, C.E. and Dobson, A.P. 2015. Bats as “special” reservoirs for emerging zoonotic pathogens. Trends Microbiol. 23(3), 172–180. Brooks, F., Wood, A.R., Thomson, J., Deane, D., Everest, D.J. and McInnes, C.J. 2014. Preliminary characterisation of Pentlands paramyxovirus -1, -2 and -3, three new paramyxoviruses of rodents. Vet. Microbiol. 170(3–4), 391–397. Brownstein, D.G., Smith, A. and Johnson, E. 1981. Sendai virus infection in genetically resistant and susceptible mice. Am. J. Pathol. 105(2), 156–163. Brownstein, D.G. 1985. Sendai virus infection, lung, mouse and rat. In Respiratory system: monographs on pathology of laboratory animals. Eds., Jones, T.C., Mohr, U. and R.D. Hunt. Heidelberg, Germany: Springer, pp: 195–203. Campbell, R.W., Carley, J.G., Doherty, R.L., Domrow, R., Filippich, C., Gorman, B.M. and Karabatsos, N. 1977. Mossman virus, a paramyxovirus of rodents isolated in Queensland. 8(11–12), 435–436. Chen, J.J., Zhang, X.A., Fan, H., Jiang, F.C., Jin, M.Z., Dai, K., Wang, N., Zhang, P.H., Li, X.K., Li, H., Shi, W., Yang, Z.C., Fang, L.Q., Zhou, H.S., Wei, Y.H. and Liu, W. 2020. Distribution and characteristics of Beilong virus among wild rodents and shrews in China. Infect. Genet. Evol. 85, 104454. Dahmana, H., Granjon, L., Diagne, C., Davoust, B., Fenollar, F. and Mediannikov, O. 2020. Rodents as hosts of pathogens and related zoonotic disease risk. Pathogens. 9(3), 202. Drexler, J.F., Corman, V.M., Müller, M.A., Maganga, G.D., Vallo, P., Binger, T., Gloza-Rausch, F., Cottontail, V.M., Rasche, A., Yordanov, S., Seebens, A., Knörnschild, M., Oppong, S., Sarkodie, Y.A., Pongombo, C., Lukashev, A.N., Schmidt-Chanasit, J., Stöcker, A., Carneiro, A.J.B., Erbar, S., Maisner, A., Fronhoffs, F., Buettner, R., Kalko, E.K.V., Kruppa, T., Franke, C.R., Kallies, R., Yandoko, E.R.N., Herrler, G., Reusken, C., Hassanin, A., Krüger, D.H., Matthee, S., Ulrich, R.G., Leroy, E.M. and Drosten, C. 2012. Bats host major mammalian paramyxoviruses. Nat. Commun. 3(1), 1–13. Faísca, P. and Desmecht, D. 2007. Sendai virus, the mouse parainfluenza type 1: a longstanding pathogen that remains up-to-date. Res. Vet. Sci. 82(1), 115–125. Han, H.J., Wen, H.L., Zhou, C.M., Chen, F.F., Luo, L.M., Liu, J.W. and Yu, X.J. 2015. Bats as reservoirs of severe emerging infectious diseases. Virus. Res. 205, 1–6. Itoh, T., Iwai, H. and Ueda, K. 1991. Comparative lung pathology of inbred strains of mice resistant and susceptible to Sendai virus infection. J. Vet. Med. Sci. 53(2), 275–279. Jack, P.J.M., Boyle, D.B., Eaton, B.T. and Wang, L.F. 2005. The complete genome sequence of J virus reveals a unique genome structure in the family Paramyxoviridae. J. Virol. 79(16), 10690–10700. Jun, M., Karabatsos, N. and Johnson, R. 1977. A new mouse paramyxovirus (J Virus). Aust. J. Exp. Biol. Med. Sci. 55(6), 645–647. Karabatsos, N., Buckley, S.M. and Ardoin, P. 1969. Nariva virus: further studies, with particular reference to its hemadsorption and hemagglutinating properties. Exp. Biol. Med. 130(3), 888–892. Kido, H., Yokogoshi, Y., Sakai, K., Tashiro, M., Kishino, Y., Fukutomi, A. and Katunuma, N. 1992. Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J. Biol. Chem. 267(19), 13573–13579. Kling, M.A. 2011. A review of respiratory system anatomy, physiology, and disease in the mouse, rat, hamster, and gerbil. Vet. Clin. North. Am. Exot. Anim. Pract. 14(2), 287–337. Lambeth, L.S., Yu, M., Anderson, D.E., Crameri, G., Eaton, B.T. and Wang, L.F. 2009. Complete genome sequence of Nariva virus, a rodent paramyxovirus. Arch. Virol. 154(2), 199–207. Lee, S.H., No, J.S., Kim, K., Budhathoki, S., Park, K., Lee, G.Y., Cho, S., Kim, B.H., Cho, S., Kim, J.W., Lee J., Cho S.H., Kim, H.C., Klein, T.A., Uhm, C.S., Kim, W.K. and Song, J.W. 2021. Novel Paju Apodemus paramyxovirus 1 and 2, harbored by Apodemus agrarius in the Republic of Korea. Virology 562, 40–49. Li, Z., Yu, M., Zhang, H., Magoffin, D.E., Jack, P.J., Hyatt, A., Wang, H.Y. and Wang, L. F. 2006. Beilong virus, a novel paramyxovirus with the largest genome of non-segmented negative-stranded RNA viruses. Virology 346(1), 219–228. MacLachlan, N.J. and Dubovi, E.J. 2017. Paramyxoviridae and Pneumoviridae. In Fenner’s veterinary virology. New York, NY: Elsevier, pp: 327–356. MacLachlan, N.J., Dubovi, E.J. and Fenner, F. 2011. Paramyxoviridae. In Fenner’s veterinary virology. New York, NY: Elsevier, pp: 299–325. Miller, P.J., Boyle, D.B., Eaton, B.T. and Wang, L.F. 2003. Full-length genome sequence of Mossman virus, a novel paramyxovirus isolated from rodents in Australia. Virology 317(2), 330–344. Natasha, J.A., Yasmin, A.R., Siti-Maisarah, A.M., Nur-Anis, Z., Tharshaini, M., Arshad, S.S., Ayuni, W.N., Mohammed, M.N. and Nur-Fazila, S.H. 2022. Screening of West Nile Virus, Herpesvirus, and Parvovirus in Rattus spp. in Klang Valley, Malaysia. Pertanika J. Trop. Agric. Sci. 45(4), 1113–1124. Nicklas, W., Bleich, A. and Mähler, M. 2012. Viral infections of laboratory mice. In The laboratory mouse. Ed., Hedrich, H.J. Academic Press, pp: 427–480. Normile, D. 2008. Rinderpest: driven to extinction. Science 319(5870), 1606–1609. Parker, J.C. and Ritcher, C.B. 1982. Viral diseases of the respiratory system. In The mouse in biomedical research. Eds., Foster, H.L., Small, J.D. and Fox, J.G. New York, NY: Academic Press, pp: 110–158. Parker, J.C., Whiteman, M.D. and Richter, C.B. 1978. Susceptibility of inbred and outbred mouse strains to Sendai virus and prevalence of infection in laboratory rodents. Infect. Immun. 19(1), 123–130. Percy, D.H., Auger, D.C. and Croy, B.A. 1994. Signs and lesions of experimental Sendai virus infection in two genetically distinct strains of SCID/eige mice. Vet. Pathol. 31(1), 67–73. Rima, B., Balkema-Buschmann, A., Dundon, W., Duprex, P., Easton, A., Fouchier, R., Kurath, G., Lamb, R., Lee, B., Rota, P., Wang, L. and Ictv Report Consortium. 2019. ICTV virus taxonomy profile: Paramyxoviridae. J. Gen. Virol. 100(12), 1593–1594. Rothenburger, J.L., Himsworth, C.G., Clifford, C.B., Ellis, J., Treuting, P.M. and Leighton, F.A. 2015. Respiratory pathology and pathogens in wild urban rats (Rattus norvegicus and Rattus rattus). Vet. Pathol. 52(6), 1210–1219. Samal, S.K. 2008. Paramyxoviruses of animals. Encycl. Virol. 40–47. Sasaki, M., Muleya, W., Ishii, A., Orba, Y., Hang’ombe, B.M., Mweene, A.S., Moonga, L., Thomas, Y., Kimura, T. and Sawa, H. 2014. Molecular epidemiology of paramyxoviruses in Zambian wild rodents and shrews. J. Gen. Virol. 95(2), 325–330. Sorden, S.D. and Castleman, W.L. 1991. Brown Norway rats are high responders to bronchiolitis, pneumonia, and bronchiolar mastocytosis induced by parainfluenza virus. Exp. Lung. Res. 17(6), 1025–1045. Tashiro, M., Yokogoshi, Y., Tobita, K., Seto, J.T., Rott, R. and Kido, H. 1992. Tryptase Clara, an activating protease for Sendai virus in rat lungs, is involved in pneumopathogenicity. J. Virol. 66(12), 7211–7216. Thibault, P.A., Watkinson, R.E., Moreira-Soto, A., Drexler, J.F. and Lee, B. 2017. Zoonotic potential of emerging paramyxoviruses. Adv. Virus. Res. 98, 1–55. Tikasingh, E.S., Jonkers, A.H., Spence, L. and Aitken, T.H. 1996. Nariva virus, a hitherto undescribed agent isolated from the Trinidadian rat, Zygodontomys b. brevicauda (J. A. Allen & Chapman). Am. J. Trop. Med. Hyg. 15(2), 235–238. Vanmechelen, B., Bletsa, M., Laenen, L., Lopes, A.R., Vergote, V., Beller, L., Deboutte, W., Korva, M., Avšič Županc, T., Goüy de Bellocq, J., Gryseels, S., Leirs, H., Lemey, P., Vrancken, B. and Maes, P. 2018. Discovery and genome characterization of three new Jeilongviruses, a lineage of paramyxoviruses characterized by their unique membrane proteins. BMC. Genomics. 19(1), 617. Walder, R. 1971. Electron microscopic evidence of Nariva virus structure. J. Med. Microbiol. 11(2), 123–128. Wang, L.F., Mackenzie, J.S. and Eaton, B.T. 2008. Disease outbreaks caused by emerging paramyxoviruses of bat origin. Emerg. Infect. Asia. 2008, 193–208. Ward, J.M., Houchens, D.P., Collins, M.J., Young, D.M. and Reagan, R.L. 1976. Naturally-occurring Sendai virus infection of athymic nude mice. Vet. Pathol. 13(1), 36–46. Wilkinson, D.A., Mélade, J., Dietrich, M., Ramasindrazana, B., Soarimalala, V., Lagadec, E., Le Minter, G., Tortosa, P., Heraud, J.M., de Lamballerie, X., and Goodman, S.M., Dellagi, K. and Pascalis, H. 2014. Highly diverse morbillivirus-related paramyxoviruses in wild fauna of the southwestern Indian Ocean Islands: evidence of exchange between introduced and endemic small mammals. J. Virol. 88(15), 8268–8277. Woo, P.C.Y., Lau, S.K.P., Wong, B.H.L., Wong, A.Y.P., Poon, R.W.S. and Yuen, K.Y. 2011. Complete genome sequence of a novel Paramyxovirus, Tailam virus, discovered in Sikkim rats. J. Virol. 85(24), 13473–13474. Woo, P.C.Y., Lau, S.K.P., Wong, B.H.L., Wu, Y., Lam, C.S.F. and Yuen, K.Y. 2012. Novel variant of Beilong Paramyxovirus in rats, China. Emerg. Infect. Dis. 18(6), 1022–1024. Woo, P.C.Y., Wong, A.Y.P., Wong, B.H.L., Lam, C.S.F., Fan, R.Y.Y., Lau, S.K.P. and Yuen, K.Y. 2016. Comparative genome and evolutionary analysis of naturally occurring Beilong virus in brown and black rats. Infect. Genet. Evol. 45, 311–319. Wu, Z., Yang, L., Yang, F., Ren, X., Jiang, J., Dong, J., Sun, L., Zhu, Y., Zhou, H. and Jin, Q. 2014. Novel henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012. Emerg. Infect. Dis. 20(6), 1064–1066. Zeltina, A., Bowden, T.A. and Lee, B. 2016. Emerging Paramyxoviruses: receptor tropism and zoonotic potential. PLOS. Pathog. 12(2), e1005390. | ||
| How to Cite this Article |
| Pubmed Style Mohd-qawiem F, Yasmin AR, Arshad SS, Norfitriah MS, Nur-fazila SH. Paramyxoviruses in Rodents: A Review. Open Vet. J.. 2022; 12(6): 877-887. doi:10.5455/OVJ.2022.v12.i6.14 Web Style Mohd-qawiem F, Yasmin AR, Arshad SS, Norfitriah MS, Nur-fazila SH. Paramyxoviruses in Rodents: A Review. https://www.openveterinaryjournal.com/?mno=24959 [Access: January 25, 2026]. doi:10.5455/OVJ.2022.v12.i6.14 AMA (American Medical Association) Style Mohd-qawiem F, Yasmin AR, Arshad SS, Norfitriah MS, Nur-fazila SH. Paramyxoviruses in Rodents: A Review. Open Vet. J.. 2022; 12(6): 877-887. doi:10.5455/OVJ.2022.v12.i6.14 Vancouver/ICMJE Style Mohd-qawiem F, Yasmin AR, Arshad SS, Norfitriah MS, Nur-fazila SH. Paramyxoviruses in Rodents: A Review. Open Vet. J.. (2022), [cited January 25, 2026]; 12(6): 877-887. doi:10.5455/OVJ.2022.v12.i6.14 Harvard Style Mohd-qawiem, F., Yasmin, . A. R., Arshad, . S. S., Norfitriah, . M. S. & Nur-fazila, . S. H. (2022) Paramyxoviruses in Rodents: A Review. Open Vet. J., 12 (6), 877-887. doi:10.5455/OVJ.2022.v12.i6.14 Turabian Style Mohd-qawiem, Firdaus, Abd Rahaman Yasmin, Siti Suri Arshad, Mohamed Sohaimi Norfitriah, and Saulol Hamid Nur-fazila. 2022. Paramyxoviruses in Rodents: A Review. Open Veterinary Journal, 12 (6), 877-887. doi:10.5455/OVJ.2022.v12.i6.14 Chicago Style Mohd-qawiem, Firdaus, Abd Rahaman Yasmin, Siti Suri Arshad, Mohamed Sohaimi Norfitriah, and Saulol Hamid Nur-fazila. "Paramyxoviruses in Rodents: A Review." Open Veterinary Journal 12 (2022), 877-887. doi:10.5455/OVJ.2022.v12.i6.14 MLA (The Modern Language Association) Style Mohd-qawiem, Firdaus, Abd Rahaman Yasmin, Siti Suri Arshad, Mohamed Sohaimi Norfitriah, and Saulol Hamid Nur-fazila. "Paramyxoviruses in Rodents: A Review." Open Veterinary Journal 12.6 (2022), 877-887. Print. doi:10.5455/OVJ.2022.v12.i6.14 APA (American Psychological Association) Style Mohd-qawiem, F., Yasmin, . A. R., Arshad, . S. S., Norfitriah, . M. S. & Nur-fazila, . S. H. (2022) Paramyxoviruses in Rodents: A Review. Open Veterinary Journal, 12 (6), 877-887. doi:10.5455/OVJ.2022.v12.i6.14 |