| Research Article | ||
Open Vet J. 2023; 13(10): 1277-1282 Open Veterinary Journal, (2023), Vol. 13(10): 1277–1282 Original Research Molecular genotyping of Salmonella spp. isolated from cheese samples of local stores in Al-Diwaniyah city, IraqOrooba Meteab Faja1,*, Afrah Sabeeh Mhyson2, Wisam Reheem Atiyah1, Basima Jasim Mohammed1 and Azal Adnan11Department of Public Health, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah City, Iraq 2Department of Conservative Treatment, College of Dentistry, University of Al-Qadisiyah, Al-Diwaniyah City, Iraq *Corresponding Author: Orooba Meteab Faja. Department of Public Health, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah City, Iraq. Email: orooba.faja [at] qu.edu.iq Submitted: 16/05/2023 Accepted: 11/09/2023 Published: 31/10/2023 © 2023 Open Veterinary Journal
AbstractBackground: Food safety is an important subject that the global cheese industry increases awareness of. This urges these economic sectors to elevate the level of research to minimize cheese contamination with pathogenic bacteria, such as Salmonella. Aim: Based on these merits, this study was conducted to genotype Salmonella spp. isolated from cheese samples of local stores in Al-Diwaniyah City, Iraq. Methods: The study used 41 samples of local fresh unsalted white cheese in a selective-growth-based isolation of Salmonella. These isolates were confirmed utilizing a slide-agglutination (SA) test and VITEK® 2 system (V2S). Then, the isolates were subjected to conventional PCR and sequencing techniques that both targeted the 16S rRNA gene. For subtyping, the Salmonella isolates were subjected to a random amplified polymorphic DNA (RAPD)-PCR method. Results: The results of both SA and V2S revealed the presence of 14 (34.2%) isolates of Salmonella spp. in the cheese samples. The PCR confirmed 6 (42.9%) of these isolates, which further were defined with close nucleotide similarity (98.03%) and (97.88%) to different world isolates, such as Salmonella enterica subsp. Arizonae and Salmonella enterica subsp. enterica serovar Typhi, respectively. The RAPD-PCR findings showed different fragments for all the tested isolates. Conclusion: The present study indicates that the samples of the local fresh unsalted white cheese contain different Salmonella genotypes, which could be originated from different contamination sources. Keywords: Cheese, Foodborne pathogens, RAPD-PCR, Salmonella, VITEK® 2. IntroductionFoodborne diseases (FBDs) are a major cause of illness and death across all demographics and a barrier to progress in both the economic and social sectors worldwide. The World Health Organization (WHO) reported in 2010 that foodborne illness was responsible for 600 million cases. In most cases, foodborne illnesses are brought on by either pathogens, including bacterial, viral, parasitic, fungal, mycotoxin, and prion-based infections, or by environmental elements, such as contamination throughout the manufacturing, preparation, transportation, and storing stages. Bacterial pathogens are the leading cause of FBDs globally, with Salmonella, Vibrio parahaemolyticus, Listeria monocytogenes, Staphylococcus aureus, and a few others being the most common and significant (Kadariya et al., 2014; Torgerson et al., 2015; Martinović et al., 2016; Bintsis 2017; Jordan and McAuliffe 2018; Faour-Klingbeil and Todd 2020; Gallo et al., 2020; Dutta et al., 2021). Salmonella, a key bacterium linked to FBDs, is often found in raw poultry, raw milk, raw meat, raw eggs, and other products. Salmonella is responsible for anything from 200 million to over a billion infections every year, resulting in 93 million incidents of gastrointestinal tract infections and diseases and 155,000 fatalities. These food items could become contaminated during production via cross-contamination. Salmonella thrived in meat, poultry, and dairy products owing to the abundance of nutrients and water in these foods (Whiley and Ross 2015; Hung et al., 2017; Kore et al., 2017; Yeh et al., 2017; Milczarek et al., 2019). In addition, food items, including fruits and vegetables contaminated by livestock fecal microorganisms, may serve as a cultivation environment for Salmonella. Salmonella causes flu-like symptoms in humans, including nausea, vomiting, stomach discomfort, and diarrhea. There are now over 2,500 known serotypes of Salmonella, with some countries having over 200 of them, such as in China. Between 1996 and 2014, the three most prevalent serotypes documented by the CDC in the United States were S. typhimurium, S. enteritidis, and S. newport (Crump et al., 2015; Chlebicz and Śliżewska, 2018; Powell et al., 2018; Sun et al., 2021; Wessels et al., 2021). The purpose of FBD tracking is to keep an eye on food contamination and dangerous components so that we can lessen the number of sick people caused by eating contaminated food. Twenty distinct kinds of FBD surveillance systems exist now, covering event-based, indicator-based, and integrated monitoring of the whole food supply chain. Salmonella has been shown to live for a long time in low-moisture foods. Bacterial pathogens may spread during cheese production, ripening, and storage as a result of direct or indirect contamination circumstances that take place in commercial, household, and retail settings. Although raw milk is generally thought to be the most common cause of cheese contamination, cross-contamination during manufacturing has been linked to the likelihood of bacteria building biofilms and surviving on food surfaces (André et al., 2008; Kousta et al., 2010; Podolak et al., 2010; Tiwari et al., 2014; Ford et al., 2015; Schön et al., 2016; Jordan et al., 2018; Chen et al., 2022). Food safety is an important subject that the global cheese industry increases awareness of. This urges these economic sectors to elevate the level of research to minimize cheese contamination with pathogenic bacteria, such as Salmonella. Based on these merits, this study was conducted to genotype Salmonella spp. isolated from cheese samples of local stores in Al-Diwaniyah City, Iraq. Materials and MethodsBacterial isolation and identificationA total of 41 samples of local Fresh unsalted white cheese were collected randomly from local stores in Al-Diwaniyah City, Iraq, during the period from October 2022 to February 2023. The samples were directly cool-transferred to the Laboratory of Public Health, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Diwaniyah City, and analyzed immediately without storage. In brief, cheese samples at 25 g/each were 225 ml-buffered peptone-suspended and incubated at 37˚C for 24 hours for pre-enrichment cultivation. Later, 1 ml of the pre-enrichment broth was utilized for selective enrichment cultivation in 10 ml tetrathionate broth and incubated at 42˚C for 24 hours. After that, 1 loop-full of the tetrathionate broth was placed onto Salmonella shigella agar and Brilliant Green Agar and incubated at 37˚C for 24 hours. Following that, the suspected Salmonella colonies were transferred to tubes that contained Kligler and Urea agar base and incubated at 37˚C for 24 hours. SA and V2S tests were performed. Molecular identification of Salmonella isolatesDNA extraction The DNA of the bacterial isolates was performed using the Wizard Genomic DNA Purification Kit (Promega, USA). The protocol of the kit was employed to complete the process. In short, 1 ml of the BHI broth (2 minutes–13,000 rpm centrifugation) pellet was used as a starting material. The resulting DNA was measured for its quality and quantity using a NanoDrop. The DNA was −20˚C-stored until further analyses. Conventional PCR The PCR was conducted using “TACGGYTACCTTGTT-ACGACTT” and “AGAGTTTGATCMTGGCTCAG” primers. At 25 µl of the total volume of the PCR reaction, the following components were inserted as: 12.5 µl (1×) master mix, 1 µl (1 μM) of each primer, 3 µl DNA, 7.5 µl water for molecular use. The following reads for a thermocycler were used; initial denaturation (denaturation, annealing, and extension) and final extension at 95˚C, (95˚C, 60˚C, and 72˚C), and 72˚C, respectively, for 5 minutes (30, 30, and 60 seconds), and 7 minutes, respectively, under 1, 30, and 1 cycle, respectively. The PCR-1%-agarose gel electrophoresis was done as described by Green and Sambrook (2019). The run was under 80–90 V for 60 minutes. The gel was visualized using a UV-light imager. RAPD-PCR The PCR was conducted using the OPB primer (Shekhawat et al., 2019) “CGT CTG GGA C.” At 50 µl of the total volume of the PCR reaction, the following components were inserted; 25 µl (10×) (EconoTaq® PULS GREEN 2X Master Mix (Lucigen), 0.5 µl primer, 1 µl DNA, and 23.5 µl water for molecular use. The following reads for a thermocycler were used; initial denaturation (denaturation, annealing, and extension) and final extension at 94˚C (94˚C, 35˚C, and 72˚C), and 72˚C, respectively, for 5 minutess (60, 60, and 120 seconds) and 7 minutes, respectively, under 1, 45, and 1 cycle, respectively. The PCR solution at 10 μl/each was mixed with 2 μl (6×) loading dye (Thermo Fisher Scientific, USA). The gel was visualized using a UV-light imager, and the images were analyzed by recruiting ImageJ software (version 1.8.0_112).
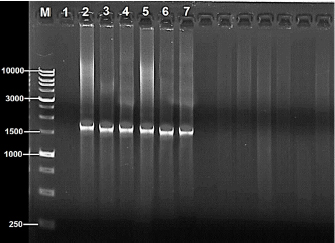
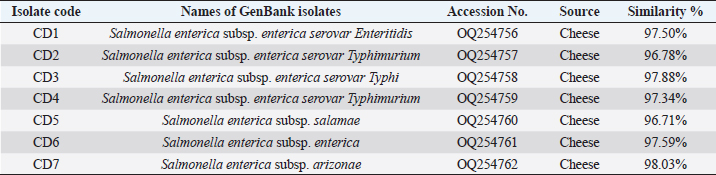
Fig. 1. PCR amplification based on 16S rRNA gene of 14 Salmonella bacterial isolates from local fresh white cheese on 1% agarose gel stained with ethidium bromide, M: 1K-ladder; 1: negative control, and 2–7: positive identification. Table 1. Comparison between the nucleotide sequences of current study Salmonella isolates with GenBank sequences (BLAST software).
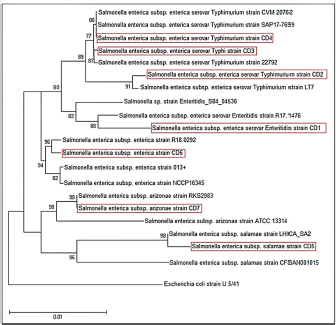
DNA sequencing The DNA from the PCR was sent to sequencing using the Sanger ABI3730XL system (Macrogen Inc., Korea). The phylogenic study and tree were computed using NCBI websites and MEGA X software. ResultsThe results of both SA and V2S revealed the presence of 14 (34.2%) isolates of Salmonella spp. in the cheese samples. The PCR confirmed 6 (42.9%) of these isolates by amplifying the 16S rRNA gene piece at 1,500 bp (Fig. 1). The sequencing defined nucleotide similarity of (98.03%) and (97.88%) with different world isolates, such as Salmonella enterica subsp. Arizonae and Salmonella enterica subsp. enterica serovar Typhi, respectively. Table 1 and Figure 2 show details about the close identity of global isolates of Salmonella spp. The RAPD-PCR findings showed different fragments for all the tested isolates.
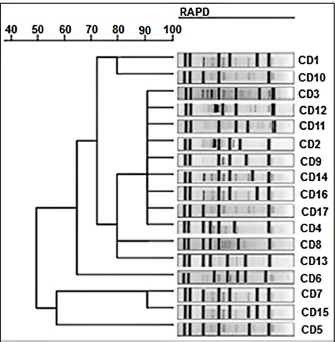
Fig. 2. Phylogenetic tree based on 16S rRNA gene of 7 Salmonella bacterial isolates from local fresh white cheese. Neighbor-joining method. The RAPD-PCR findings showed different fragments for all the tested isolates. Some isolates showed universal and distinct bands (Fig. 3). DiscussionThe present study revealed that the local Iraqi white cheese has different Salmonella isolates with different subtypes. Sanitize industrial machinery, microorganism-free technological water, and sanitary conditions during packing and storage at the distributor are all necessary for a safe and effective manufacturing process. Cheese is a fantastic medium for bacterial development because of its contents of protein, lactose, and water content. Thus, the short shelf life of local cheeses may be due in part to bacteria in milk or arise from secondary contamination. An efficient cleaning process, as well as hygienic conditions for packing and storing the product, are essential for achieving the highest possible hygiene requirements (Schön et al., 2016).
Fig. 3. RAPD-PCR-based dendrogram of subtyping for Salmonella bacterial isolates from local fresh white cheese. Average linkage unweighted group pair method with arithmetic averages (UPGMA). Inadequate sanitary practices during milking and on farms lead to the prevalence of coliform bacteria in raw milk. Furthermore, coliform bacteria may be used as a gauge of the sanitation of dairy processing facilities. The presence of coliform bacteria was identified in 45% of the examined samples in research by Lobacz and Zulewska (2021) that assessed the microbiological quality of tvarogs from 15 producers. All of the samples from the raw milk cheese tested positive for the presence of fecal bacteria, with an average starting count of 6 log cfu/g (Lobacz and Zulewska, 2021). Given that S. enteritidis may survive in the products for many months, Salmonella infection in sheep poses a genuine concern to public health. Sheep milk is often not pasteurized before cheese manufacture. Raw milk cheese was shown to be the origin of infection after a microbiological and environmental examination, as well as molecular typing of S. enteritidis isolates (Doosti et al., 2017; Napoleoni et al., 2021). Our sequencing results indicated close identity with global isolates of Salmonella spp. Many facts can play a role in introducing new base pairs to local bacterial species. One of the important factors is the travel and import of animals from endemic areas and countries. This process can bring in a novel bacterial species, and with the presence of different genetic tools, such as horizontal gene transfer, a new bacterial sequence can be produced. Then, the newly produced bacterial isolates can show close nucleotide similarity with global sequences, which can hit 98% similarity (Wang et al., 2017; Bokhary et al., 2021; Sridhar et al., 2021). All of the Salmonella spp. isolates examined were able to be fingerprinted using RAPD-PCR. Salmonella isolates clustered into seven distinct RAPD-PCR types in a dendrogram constructed using the RAPD-PCR technique. Dendrogram fingerprint analysis revealed four distinct fingerprint types among all Salmonella isolates, clustering them into seven distinct clonal groupings. The Salmonella isolates showed average genetic similarity. Some isolates revealed similar bands, which might indicate a conserved sequence between isolates. The current study results agree with those of Shekhawat et al. (2019), who identified different subtypes of four Salmonella isolates from different foods. Shekhawat et al. (2019) found that Salmonella gallinarum isolates from chickens had the most genetic diversity (Shekhawat et al., 2019). It is important to notice that detecting pathogenic bacteria in cheese samples brings high attention to public health concerns. This is true since these bacteria may carry antibiotic-resistance genes in their genomes that can be transmitted between the same bacterial species or to other bacterial species via genetic carriers, such as plasmids. Chahouri et al. (2022) studied the presence of fecal bacteria and other bacterial organisms and their antibiotic resistance pattern in marine and river ecosystems. The authors detected the presence of Salmonella spp. in their samples and were susceptible to all antibiotics tested except amoxicillin+Ac clavulanic, ampicillin, and chloramphenicol (Chahouri et al., 2022). However, this indicates that the current study isolates may carry the same pattern or with a broader range of antibiotic resistance, which makes cheese as one of the main sources of this health risk. The present study indicates that the samples of the local fresh unsalted white cheese contain different Salmonella genotypes, which could have originated from different contamination sources. AcknowledgmentsNone. Conflict of interestAll authors of the current article confirm that this work has no conflict of interest. Authors contributionsAll authors participated in the sample, Lab work, data analysis, and writing, revising, and approving the manuscript. FundingSelf-funded by the authors. Data availabilityData of the current study are available upon request. ReferencesAndré, M.C.D.P.B., Campos, M.R.H., Borges, L.J., Kipnis, A., Pimenta, F.C., and Serafini, Á.B. 2008. Comparison of Staphylococcus aureus isolates from food handlers, raw bovine milk and Minas Frescal cheese by antibiogram and pulsed-field gel electrophoresis following SmaI digestion. Food Control. 19(2), 200–207. Bintsis, T. 2017. Foodborne pathogens. AIMS Microbiol. 3(3), 529–563. Bokhary, H., Pangesti, K.N.A., Rashid, H., Abd El Ghany, M., and Hill-Cawthorne, G.A. 2021. Travel-related antimicrobial resistance: a systematic review. Trop. Med. Infect. Dis. 6(1), 11–37. Chahouri, A., Radouane, N., Yacoubi, B., Moukrim, A., & Banaoui, A. (2022). Microbiological assessment of marine and estuarine ecosystems using fecal indicator bacteria, Salmonella, Vibrio and antibiotic resistance pattern. Marine Poll. Bull. 180, 113824. Chen, L., Sun, L., Zhang, R., Liao, N., Qi, X., and Chen, J. 2022. Surveillance for foodborne disease outbreaks in Zhejiang Province, China, 2015–2020. BMC Public Health. 22(1), 135–143. Chlebicz, A., and Śliżewska, K. 2018. Campylobacteriosis, salmonellosis, yersiniosis, and listeriosis as zoonotic foodborne diseases: a review. Int. J. Environ. Res. Public Health. 15(5), 863–890. Crump, J. A., Sjölund-Karlsson, M., Gordon, M. A., and Parry, C. M. 2015. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 28(4), 901–937. Doosti, A., Doosti, E., Rahimi, E., and Ghasemi-Dehkordi, P. 2017. Frequency of antimicrobial-resistant genes in Salmonella enteritidis isolated from traditional and industrial Iranian white cheeses. Proc. Natl. Acad. Sci. India Sect. B. Biol. Sci. 87(1), 73–80. Dutta, D., Kaushik, A., Kumar, D., and Bag, S. 2021. Foodborne pathogenic vibrios: antimicrobial resistance. Front. Microbiol. 12(6), 638331–638340. Faour-Klingbeil, D., and Todd, E.C.D. 2020. Prevention and control of foodborne diseases in Middle-East North African Countries: review of national control systems. Int. J. Environ. Res. Public Health. 17(1), 70–92. Ford, L., Miller, M., Cawthorne, A., Fearnley, E., and Kirk, M. 2015. Approaches to the surveillance of foodborne disease: a review of the evidence. Foodborne Pathog. Dis. 12(12), 927–936. Gallo, M., Ferrara, L., Calogero, A., Montesano, D., and Naviglio, D. 2020. Relationships between food and diseases: What to know to ensure food safety. Food Res. Int. 137(6), 109414–109419. Green, M.R., and Sambrook, J. 2019. Analysis of DNA by agarose gel electrophoresis. Cold Spring Harb. Protoc. 2019(1), 6–15. Hung, Y.T., Lay, C.J., Wang, C.L., and Koo, M. 2017. Characteristics of nontyphoidal Salmonella gastroenteritis in Taiwanese children: A 9-year period retrospective medical record review. J. Infect. Public Health. 10(5), 518–521. Jordan, K., Hunt, K., Lourenco, A., and Pennone, V. 2018. Listeria monocytogenes in the food processing environment. Curr. Clin. Microbiol. Rep. 5(2), 106–119. Jordan, K., and McAuliffe, O. 2018. Listeria monocytogenes in foods. Adv. Food Nutr. Res. 86(4), 181–213. Kadariya, J., Smith, T.C., and Thapaliya, D. 2014. Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in Public Health. Biomed. Res. Int. 2014(4), 827965–827973. Kore, K., Asrade, B., Demissie, K., and Aragaw, K. 2017. Characterization of Salmonella isolated from apparently healthy slaughtered cattle and retail beef in Hawassa, southern Ethiopia. Prev. Vet. Med. 147(11), 11–16. Kousta, M., Mataragas, M., Skandamis, P., and Drosinos, E.H. 2010. Prevalence and sources of cheese contamination with pathogens at farm and processing levels. Food Control. 21(6), 805–815. Lobacz, A., and Zulewska, J. 2021. Fate of Salmonella spp. in the fresh soft raw milk cheese during storage at different temperatures. Microorganisms 9(5), 938–951. Milczarek, M., Sadkowska-Todys, M., Czarkowski, M.P. and Kitowska, W. 2019. Salmonellosis in Poland in 2017. Prz. Epidemiol. 73(4), 463–477. Martinović, T., Andjelković, U., Gajdošik, M.Š., Rešetar, D., and Josić, D. 2016. Foodborne pathogens and their toxins. J. Proteomics. 147(9), 226–235. Napoleoni, M., Villa, L., Barco, L., Busani, L., Cibin, V., Lucarelli, C., Tiengo, A., Dionisi, A. M., Conti, F., Nunes, F.R.D.S., Tantucci, L., Staffolani, M., Silenzi, V., Fraticelli, R., Morandi, B., Blasi, G., Rocchegiani, E., and Fisichella, S. 2021. A strong evidence outbreak of Salmonella enteritidis in Central Italy linked to the consumption of contaminated raw sheep milk cheese. Microorganisms. 9(12), 2472. Podolak, R., Enache, E., Stone, W., Black, D.G., and Elliott, P.H. 2010. Sources and risk factors for contamination, survival, persistence, and heat resistance of Salmonella in low-moisture foods. J. Food Prot. 73(10), 1919–1936. Powell, M.R., Crim, S.M., Hoekstra, R.M., Williams, M.S., and Gu, W. 2018. Temporal patterns in principal Salmonella serotypes in the USA; 1996–2014. Epidemiol. Infect. 146(4), 437–441. Schön, K., Schornsteiner, E., Dzieciol, M., Wagner, M., Müller, M., and Schmitz-Esser, S. 2016. Microbial communities in dairy processing environment floor-drains are dominated by product-associated bacteria and yeasts. Food Control. 70(12), 210–215. Shekhawat, S.S., Gaurav, A., Joseph, B., Kumar, H., and Kumar, N. 2019. Random amplified polymorphic DNA-based molecular heterogeneity analysis of Salmonella enterica isolates from foods of animal origin. Vet. World. 12(1), 146–154. Sridhar, S., Turbett, S.E., Harris, J.B., and LaRocque, R.C. 2021. Antimicrobial-resistant bacteria in international travelers. Curr. Opin. Infect. Dis. 34(5), 423–431. Sun, T., Liu, Y., Qin, X., Aspridou, Z., Zheng, J., Wang, X., Li, Z., and Dong, Q. 2021. The prevalence and epidemiology of Salmonella in retail raw poultry meat in China: a systematic review and meta-analysis. Foods. 10(11), 2757–2767. Tiwari, U., Walsh, D., Rivas, L., Jordan, K., and Duffy, G. 2014. Modelling the interaction of storage temperature, pH, and water activity on the growth behaviour of Listeria monocytogenes in raw and pasteurised semi-soft rind washed milk cheese during storage following ripening. Food Control. 42(8), 248–256. Torgerson, P.R., Devleesschauwer, B., Praet, N., Speybroeck, N., Willingham, A.L., Kasuga, F., Rokni, M.B., Zhou, X.N., Fèvre, E.M., Sripa, B., Gargouri, N., Fürst, T., Budke, C.M., Carabin, H., Kirk, M.D., Angulo, F.J., Havelaar, A., and de Silva, N. 2015. World Health Organization estimates of the global and regional disease burden of 11 foodborne parasitic diseases, 2010: a data synthesis. PLoS Med. 12(12), e1001920–e1001941. Wang, J., Ma, Z.B., Zeng, Z.L., Yang, X.W., Huang, Y., and Liu, J.H. 2017. The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zool. Res. 38(2), 55–80. Wessels, K., Rip, D., and Gouws, P. 2021. Salmonella in chicken meat: consumption, outbreaks, characteristics, current control methods and the potential of bacteriophage use. Foods. 10(8), 1742–1761. Whiley, H., and Ross, K. 2015. Salmonella and eggs: from production to plate. Int. J. Environ. Res. Public Health. 12(3), 2543–2556. Yeh, Y., Purushothaman, P., Gupta, N., Ragnone, M., Verma, S.C., and de Mello, A.S. 2017. Bacteriophage application on red meats and poultry: effects on Salmonella population in final ground products. Meat Sci. 127(5), 30–34. | ||
| How to Cite this Article |
| Pubmed Style Faja OM, Mhyson AS, Atiyah WR, Mohammed BJ, Adnan A. Molecular genotyping of Salmonella spp isolated from cheese samples of local stores in Al-Diwaniyah city, Iraq. Open Vet J. 2023; 13(10): 1277-1282. doi:10.5455/OVJ.2023.v13.i10.6 Web Style Faja OM, Mhyson AS, Atiyah WR, Mohammed BJ, Adnan A. Molecular genotyping of Salmonella spp isolated from cheese samples of local stores in Al-Diwaniyah city, Iraq. https://www.openveterinaryjournal.com/?mno=153433 [Access: July 01, 2025]. doi:10.5455/OVJ.2023.v13.i10.6 AMA (American Medical Association) Style Faja OM, Mhyson AS, Atiyah WR, Mohammed BJ, Adnan A. Molecular genotyping of Salmonella spp isolated from cheese samples of local stores in Al-Diwaniyah city, Iraq. Open Vet J. 2023; 13(10): 1277-1282. doi:10.5455/OVJ.2023.v13.i10.6 Vancouver/ICMJE Style Faja OM, Mhyson AS, Atiyah WR, Mohammed BJ, Adnan A. Molecular genotyping of Salmonella spp isolated from cheese samples of local stores in Al-Diwaniyah city, Iraq. Open Vet J. (2023), [cited July 01, 2025]; 13(10): 1277-1282. doi:10.5455/OVJ.2023.v13.i10.6 Harvard Style Faja, O. M., Mhyson, . A. S., Atiyah, . W. R., Mohammed, . B. J. & Adnan, . A. (2023) Molecular genotyping of Salmonella spp isolated from cheese samples of local stores in Al-Diwaniyah city, Iraq. Open Vet J, 13 (10), 1277-1282. doi:10.5455/OVJ.2023.v13.i10.6 Turabian Style Faja, Orooba Meteab, Afrah Sabeeh Mhyson, Wisam Reheem Atiyah, Basima Jasim Mohammed, and Azal Adnan. 2023. Molecular genotyping of Salmonella spp isolated from cheese samples of local stores in Al-Diwaniyah city, Iraq. Open Veterinary Journal, 13 (10), 1277-1282. doi:10.5455/OVJ.2023.v13.i10.6 Chicago Style Faja, Orooba Meteab, Afrah Sabeeh Mhyson, Wisam Reheem Atiyah, Basima Jasim Mohammed, and Azal Adnan. "Molecular genotyping of Salmonella spp isolated from cheese samples of local stores in Al-Diwaniyah city, Iraq." Open Veterinary Journal 13 (2023), 1277-1282. doi:10.5455/OVJ.2023.v13.i10.6 MLA (The Modern Language Association) Style Faja, Orooba Meteab, Afrah Sabeeh Mhyson, Wisam Reheem Atiyah, Basima Jasim Mohammed, and Azal Adnan. "Molecular genotyping of Salmonella spp isolated from cheese samples of local stores in Al-Diwaniyah city, Iraq." Open Veterinary Journal 13.10 (2023), 1277-1282. Print. doi:10.5455/OVJ.2023.v13.i10.6 APA (American Psychological Association) Style Faja, O. M., Mhyson, . A. S., Atiyah, . W. R., Mohammed, . B. J. & Adnan, . A. (2023) Molecular genotyping of Salmonella spp isolated from cheese samples of local stores in Al-Diwaniyah city, Iraq. Open Veterinary Journal, 13 (10), 1277-1282. doi:10.5455/OVJ.2023.v13.i10.6 |