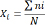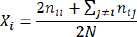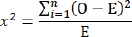| Research Article | ||
Open Vet. J.. 2024; 14(12): 3248-3256 Open Veterinary Journal, (2024), Vol. 14(12): 3248-3256 Research Article Body weight and measurement traits of Brahman cattle affected by pleomorphic adenoma gene 1 variationsBaharudin Trilaksono1†, Amira Fathin Rodhiyah1†, Yuli Yanti1, Joko Riyanto1, Hendra Kurniawan2, Muhammad Imron2, Delly Nista2, Yumoko Ginto2, Pita Sudrajad3 and Muhammad Cahyadi1*1Department of Animal Science, Universitas Sebelas Maret, Surakarta, Indonesia 2Balai Pembibitan Ternak Unggul dan Hijauan Pakan Ternak (BPTU-HPT) Sembawa, Ministry of Agriculture, Sumatera Selatan, Indonesia 3Research Center for Animal Husbandry, National Research and Innovation Agency—Indonesia (BRIN), Cibinong, Indonesia †These authors contributed equally to this work *Corresponding Author: Muhammad Cahyadi. Department of Animal Science, Universitas Sebelas Maret, Surakarta, Indonesia. Email: mcahyadi [at] staff.uns.ac.id Submitted: 15/08/2024 Accepted: 25/11/2024 Published: 31/12/2024 © 2024 Open Veterinary Journal
AbstractBackground: Pleomorphic adenoma gene 1 (PLAG1) is a zinc finger transcription factor and is widely known to play an important role in regulating growth traits in bovine. Aim: This study aimed to evaluate the association of PLAG1 polymorphisms with body weight and measurement traits of Brahman cattle. Methods: A total of 57 blood samples of Brahman cattle and their body weight and measurement traits have been collected. Moreover, the polymerase chain reaction-restriction fragment length polymorphism method was used to determine the genotypes of the three polymorphisms in PLAG1, namely g.48308C>T, g.32212 (19 bp indel), and g.45233 T>C. A mix model effect was performed to evaluate the association of PLAG1 with body weight and measurement traits, while the Tukey HSD test was used to compare the means among genotypes. Results: Genotype variations were observed for the g.48308C>T single nucleotide polymorphism (SNP) and 19 bp indel. In detail, the CC and CT genotypes of g.48308C>T SNP were identified and all possible genotypes of 19 bp indel were also identified. On the other hand, g.45233T>C SNP showed no variation. The statistical analysis revealed that SNP g.48308C>T and 19 bp indel had a significant effect on body weight and chest girth (p < 0.05). Both CC genotypes of g.48308C>T SNP and DD genotype of 19 bp indel of the PLAG1 were favorable for those two traits. Conclusion: Based on these results, the SNP g.48308C>T and 19 bp indel in the PLAG1 could be a candidate marker for body weight and chest girth in the Brahman cattle population at the Center for Superior Animal Breeding and Forage (BPTU-HPT) Sembawa. Keywords: Association study, Body weight, Brahman cattle, Chest girth, PLAG1 variations. IntroductionThe beef cattle farming business in Indonesia is largely dominated by small-scale farmers who use traditional maintenance management and own between 5 and 10 heads (Indrayani and Andri, 2018). In this context, cattle are often kept on the farm by grazing, penning, and moving around residential areas (Rusdiana and Praharani, 2018). Despite the potential of the business, farmers typically face various challenges, such as difficulty in obtaining superior breeds (Maskur et al., 2023). In Indonesia, the development of beef cattle farming is primarily driven by the people’s awareness and high demand for beef as nutritious food (Handayanta et al., 2016). However, beef cattle farming in the country has not been able to meet domestic animal food needs (Rusdiana and Praharani, 2018). In line with previous studies, several breeds of beef cattle have been identified in Indonesia, including Brahman. Novita et al. (2019) reported that Brahman cattle are a descendant of Zebu cattle (Bos indicus), which grow quickly in tropical climates, such as the United States, Australia, New Zealand, and Indonesia. Research has emphasized the superiority of Brahman cattle in terms of resistance to heat stress, ticks, and overall growth performance compared to common local cattle breeds, making them promising candidates for further breeding and development programs in Indonesia (Priyadi et al., 2017). In addition, this breed has the potential to be developed for increasing food production, specifically beef in Indonesia (Sholikah et al., 2018). Building on this idea, Suhada et al. (2009) revealed that cattle productivity is influenced by two factors, namely external factors such as environment and genetics factor. External factors are typically contemporary or changing and cannot be inherited, while genetic factors are fixed, do not change throughout life except when subjected to mutations, and can be passed on to offspring (Suhada et al., 2009). Hikmawaty et al. (2018) also stated that genetic factors can influence morphometric differences between individual cows, indicating their role in the success of the livestock industry (Bilyaro et al., 2023). Pleomorphic adenoma gene 1 (PLAG1) is a zinc finger transcription factor gene located on chromosome 14 in cattle (BTA14), which influences body size and reproductive traits (Sukaryo et al., 2022). According to Hartati et al. (2015), it has the highest expression effect on growth traits. The gene also encodes a transcription factor that is widely expressed during fetal development and is inherited at birth. In addition, it can also cause the modulation of the like growth factor (IGF1), IGF1R, IGF2, GH1, and GHR networks, which are known as key actors in the growth pathway (Pereira et al., 2016). Genetic variations, such as single nucleotide polymorphism (SNP) and insertion-deletion (indel) in the PLAG1 can affect body size in cattle. Previous studies have shown that PLAG1 has a significant correlation with birth weight (Fathira et al., 2022), body length in Bali cattle (Cahyadi et al., 2022), body height and chest girth in Yunling cattle (Zhou et al., 2019), weight in beef cattle (Fortes et al., 2013; Utsunomiya et al., 2017), and carcass weight (Song et al., 2016). Zhang et al. (2020) also reported its role in the production of molecular markers related to growth traits. Body weight and measurement traits are two easily observed phenotypic traits representing the growth traits of cattle (Kamprasert et al., 2019; Bramastya et al., 2022). They are directly associated with body weight gain (Cominotte et al., 2020) and are widely used as predictors of production traits in beef cattle (Naserkheil et al., 2020). Moreover, their estimated heritabilities are moderate to high in Brahman cattle (Kamprasert et al., 2019) indicating that genetic factor plays a big role in determining these traits. Despite the potential, there is limited information regarding the association between PLAG1 and body weight and measurement traits in Brahman cattle, specifically in Indonesia. Therefore, this study aimed to evaluate the association of PLAG1 with body weight and measurement traits in Brahman cattle at the Center for Superior Animal Breeding and Forage (BPTU-HPT) Sembawa. Materials and MethodsPopulation characteristicsIn this study, blood samples were obtained from 57 healthy Brahman cattle at the BPTU-HPT Sembawa, with an average age of 2 years. Brahman cattle were kept using a semi-intensive rearing system. The system is characterized by the consumption of forage through grazing, while concentrate and ad libitum drinking water were provided in the pen. All animals received 3 kg of standard concentrate per day and consumed 30 kg of grass per day. The Brahman cattle used in this study should have complete records of body weight at 2 years and body measurement traits including body length, withers height, chest girth, and waist height (Fig. 1). Blood sample collection and DNA extractionA total of 5 ml of blood samples were taken from the veins using a venoject in a 10 ml vacutainer tube containing EDTA. These samples were stored in a cooler at 0°C and a freezer at −21°C for DNA extraction. Subsequently, DNA extraction was carried out using a DNA Extraction Kit for blood (Geneaid Biotech Ltd., Taiwan) following the protocol provided by the manufacturer. PLAG1 gene amplification and genotypingAmplification of PLAG1 fragment was carried out to identify three PLAG1 variations, namely g.48308C>T, g.32212 (19 bp indel), and g.45233 T>C. The primer pairs used in this study were presented in Table 1. The PCR reaction consisted of 12.5 µl master mix, 9.5 µl nuclease-free water, 1 µl primer, and 1 µl DNA template. All materials were mixed in a PCR tube with a total volume of 25 µl. In addition, amplification was carried out in sequential stages starting from pre-denaturation at 94°C for 5 minutes, denaturation at 94°C for 30 seconds and 35 cycles, annealing with temperature and time as in Table 1, extension at 72°C for 30 seconds, and final extension at 72°C for 10 minutes. PCR products were run using 1.5% agarose gel stained with ethidium bromide and then electrophoresed using an electrophoresis machine (Mupid-EX®, Japan) for 30 minutes at 100V. Standard DNA band size was determined using a 100 bp marker ladder, followed by visualization with the Gel Documentation System (Glite UV, Pacific Image, Taiwan). Genotyping was carried out by observing PCR products DNA bands to detect three PLAG1 polymorphisms. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) was then carried out to detect the presence of cutting bands on the agarose gel. Additionally, RFLPs were performed according to the Fast Digest SacII protocol for g.48308C>T SNP and Fast Digest BclI for g.45233T>C SNP. On the other hand, the g.32212 (19 bp indel) was identified by directly observing PCR products sizes. The products obtained were visualized under UV light using the Gel Documentation System. Data analysisGenotype frequency was calculated using the formula proposed by Nei and Kumar (2000) as follows:
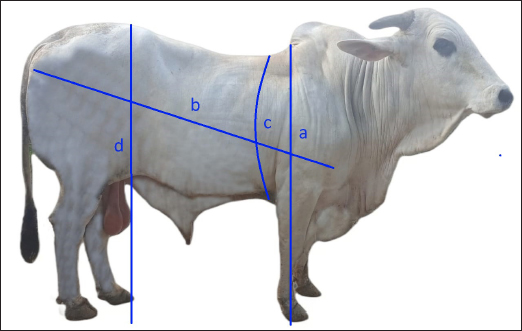
Fig. 1. Body measurement of Brahman cattle in the BPTU-HPT Sembawa. Line a represents withers height; b is body length; c is chest girth; and d is waist height. Table 1. Primer pairs of the PLAG1.
Xi is the frequency of genotype i, ni is the total of individuals of genotype i, and N is the total of samples observed. Allele frequency was also calculated using the Nei and Kumar (2000) formula as follows:
Xi is the frequency of the i-th allele, nii is the total of individuals with the AiAi genotype, nij is the total of individuals with the AiAj genotype, and N is the total of samples. Hardy-Weinberg Equilibrium (HWE) was determined based on observed and expected genotype frequencies. The chi-square goodness of fit test was used to determine the nature of the difference between observed and expected frequencies to determine whether the population was in HWE or not. The Hardy-Weinberg equation was as follows: p + q=1 p2 + 2pq + q2=1 Note: p is the frequency of allele 1 and q is the frequency of allele 2. HWE was tested with chi-square (x2) (Guo and Thomson, 1992),
X2 is the chi-square test, O is the total of observations of the ith genotype, and E is the expected total of the ith genotype.
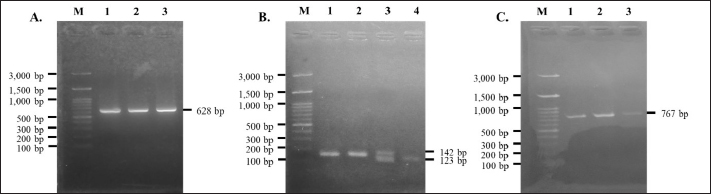
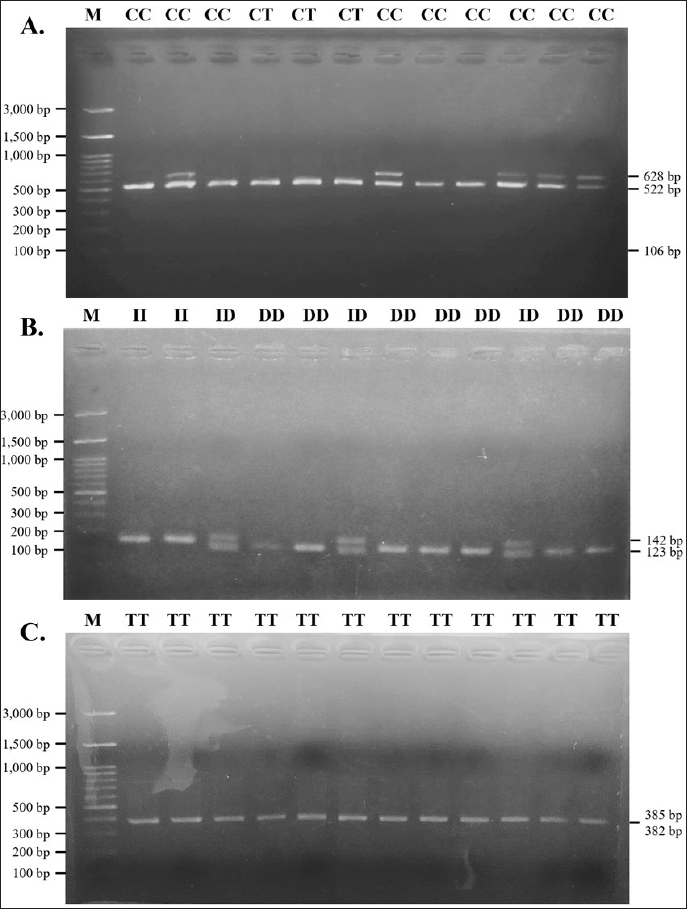
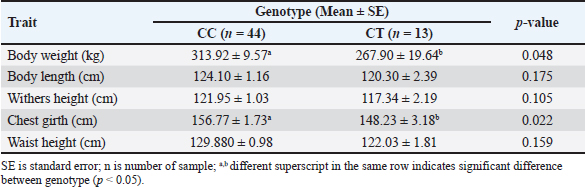
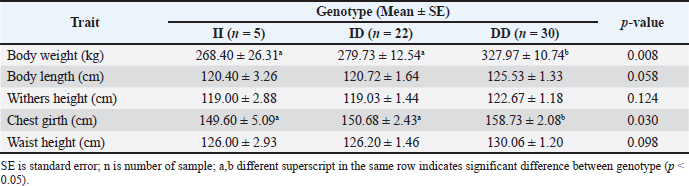
Fig. 2. Agarose gel electrophoresis of PCR products targeting three polymorphisms in PLAG1. (A) is PCR product targeting g.48308C>T SNP; (B) is PCR product targeting g.32212 (19 bp indel); (C) is PCR product targeting g.45233T>C SNP. M is 100 bp marker ladder and 1 to 4 represent individual Brahman cattle. Association of PLAG1 with body weight and body measurement traits in Brahman cattleMix effect models were used to evaluate the association of PLAG1 polymorphisms with body weight and measurement traits of Brahman cattle using MINITAB version 19.1 software. Genotype was used as a fixed effect, dam was used as a random factor, and body weight, body length, withers height, chest girth, and waist height served as response variables. Further tests were carried out on significant results with the Tukey HSD test to determine differences among genotypes. Ethical approvalAll animal procedures related to blood collections of Brahman cattle at the BPTU-HPT Sembawa were approved by the Ethical Clearance Commission for Research of the National Research and Innovation Agency (BRIN) with decree No. 81/Klirens/X/2021. ResultsAmplification and genotyping of PLAG1 in Brahman cattleAmplifications of PLAG1 targeting 19 bp indel, g.48308C>T, and g.45233T>C SNPs were successfully carried out. The results of PCR amplification of PLAG1 in this study are presented in Figure 2. An amplicon of 628 bp was identified at SNP g.48308C>T (Fig. 2A), 142/123 bp was recognized at SNP g.32212 (Fig. 2B), and 767 bp was identified at g.45233T>C SNP (Fig. 2C). Genotypes of those three variations of PLAG1 are presented in Figure 3. Genotypes and allele frequencies of PLAG1 in Brahman cattleBrahman cattle at the BPTU-HPT Sembawa were observed to have two genotypes for the g.48308C>T SNP, namely CC and CT, while the TT genotype was not observed. The CC genotype and C allele were mostly found in the Brahman cattle population. In addition, three different genotypes were observed for g.32212 19 bp indel, namely II, ID, and DD. The frequency of the DD genotype was dominant compared to others and D allele was much higher than I allele in this study. Genotypes and allele frequencies of the g.48308C>T SNP and g.32212 19 bp indel are presented in Tables 2 and 3. Association of PLAG1 with body weight and measurement traits of Brahman cattleThe results of the statistical analysis of the association between g.48308C>T SNP and 19 bp indel with body weight and measurement traits of Brahman cattle are presented in Tables 4 and 5. Statistical analysis showed that g.48308C>T SNP was significantly associated with body weight and chest girth with respective p-values of 0.048 and 0.022. Brahman cattle carrying the CC genotype had higher body weight and chest girth than the animals with the TT genotype (Table 4). Moreover, significant associations were also found between 19 bp indel variations with body weight and chest girth with DD genotype was favorable for those two traits (Table 5). DiscussionAmplification and genotyping of PLAG1 in Brahman cattleThe PLAG1 was a member of the PLAG family as a zinc finger transcription factor, which had a specific relationship with growth traits in humans and various livestock species, including cattle (Mia et al., 2022). In addition, it had variations in the coding region of the genome (allelic), which had a good relationship with stature and age at puberty in cattle (Engle and Hayes, 2022). This gene was identified as having an influence on body size and reproductive traits (Sukaryo et al., 2022). This study was conducted to determine the relationship between g.48308C>T SNP, g.32212 19 bp indel, and g.45233T>C SNP with body weight and body measurement traits of Brahman cattle. PCR-RFLP was used to determine the genotype of each Brahman cattle sample. Akmalputra et al. (2022) reported that the RFLP analysis of the PCR product from PLAG1 with SNP g.48308C>T using the SacII restriction enzyme revealed three genotypes. The CC genotype was cut into fragments of 522 bp and 106 bp, the CT genotype resulted in DNA fragments of 628, 522, and 106 bp, and the TT genotype produced a DNA fragment of 628 bp, which was resistant to digestion by the endonuclease. In addition, the results showed that the SacII enzyme had a CCGCGG cutting site. The PCR-RFLP product from PLAG1 with g.48308C>T SNP in Brahman cattle at the BPTU-HPT Sembawa in this report was polymorphic because there were two different genotypes observed, namely CC and CT. However, TT genotype in this study was not observed. The results were different compared to those of Akmalputra et al. (2022), and Sukaryo et al. (2023) that three genotypes had been found, namely CC, CT, and TT in Bali and PO cattle.
Fig. 3. Agarose gel electrophoresis of PCR-RFLP products targeting g.48308C>T SNP (A), g.32212 (19 bp indel) (B), and g.45233T>C SNP (C) of the PLAG1. M is 100 bp marker ladder. Table 2. Allele and genotype frequencies, and chi-square test of the g.48308C>T SNP.
Table 3. Allele and genotype frequencies, and chi-square test of the g.32212 (19 bp indel).
Table 4. Association of the g.48308C>T SNP and body weight and measurement traits of Brahman cattle.
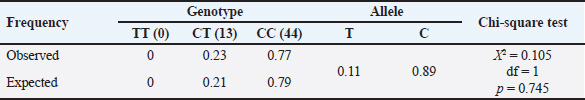
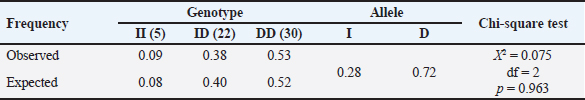
The g.32212 19 bp indel in PLAG1 was located in intron 3 and had 3 genotypes, namely homozygous II, heterozygous ID, and homozygous DD (Cahyadi et al., 2022). DNA band fragments with 142 bp and 123 bp representing g.32212 19 bp indel in this report obtained varying results (polymorphic) because three different genotypes were detected, which included genotype DD (123 bp), II (142 bp), and ID genotype which had two bands (123 and 142 bp). The results obtained by Xu et al. (2018) were similar, where three genotypes of 19 bp indel were found, namely II (142 bp), ID (142 bp and 123 bp), and DD (123 bp). However, the findings were different with Sukaryo et al. (2022) in Bali cattle, which only found a genotype, namely homozygous DD. Sukaryo et al. (2023) and Akmalputra et al. (2022) in PO cattle also showed two genotypes, namely heterozygous ID and homozygous DD, while genotype II was not observed. Another SNP, namely g.45233 T>C, was also successfully found on a 767 bp DNA fragment. The g.45233T>C SNP was a missense variant and was located in exon 4 (Cahyadi et al., 2022). This SNP did not vary or be known as monomorphic because all samples had the same genotype. The results of this study were consistent with Cahyadi et al. (2022) and Akmalputra et al. (2022) that SNP g.45233T>C did not vary in Bali and PO cattle population. Genotype and allele frequencies of PLAG1 in Brahman cattleBrahman cattle at the BPTU-HPT Sembawa were observed to have two genotypes with observed frequencies of CC and CT genotypes were 77% and 23%, respectively. The frequencies of C and T alleles were 0.89 and 0.11, respectively. These results indicated that CC genotype was more dominant in Brahman cattle. These findings were in line with a previous report conducted by Cahyadi et al. (2022) that CC genotype was more dominant in Bali cattle (Bos javanicus). Zhong et al. (2019) also reported that CC genotype was more dominant than TT genotype in Pinan, Ji’an, Qingchuan, and Xianan cattle. In terms of 19 bp indel, DD genotype was mostly found in this study with consistent with previous findings conducted by Sukaryo et al. (2023) and Akmalputra et al. (2022) who also reported that DD genotype was dominant in PO cattle. Table 5. Association of the 19 bp indel and body weight and measurement traits of Brahman cattle.
Based on the results of the chi-square test for the HWE in Brahman cattle, the significance value was greater than 0.05 (p > 0.05). However, the distribution of Brahman cattle genotypes was statistically consistent with the HWE model. A population was said to be in HWE when there was no migration and natural selection (Lachance, 2016). Allendorf and Luikart et al. (2007) stated that when the population was in HWE, the genotype and allele frequencies were constant from generation to generation because the combination of gametes occurred randomly in a fairly large population. Association of PLAG1 with body weight and measurement traits of Brahman cattleThe results indicated that genetic variation at the SNP position g.48308C>T of PLAG1 had a significant relationship with the physical characteristics of Brahman cattle. These findings were consistent with Zhong et al. (2019) who reported that g.48308C>T SNP had a significant influence on the growth and development of 5 cattle breeds in China (p < 0.05). Cattle with the CC genotype were more common in these traits and had superior growth characteristics. The results of this study were in line with Fortes et al. (2013), who found that PLAG1 influenced body weight. These results confirmed the significant role of PLAG1 in regulating the growth and physical characteristics. The 19 bp indel polymorphism in PLAG1 was body weight and chest girth. This result was the same as a previous study conducted by Zhou et al. (2019) who reported that a 19 bp indel in PLAG1 was significantly associated with chest girth in Yunling cattle. Xu et al. (2018) also reported that there was a significant association of 19 bp indel with body weight in four breeds of Chinese cattle and significant with chest girth in Xianan and Jiaxian cattle. Similar findings in different contexts demonstrated the consistent role of the 19 bp indel polymorphism in PLAG1 in body weight and measurement traits of cattle. The PLAG1 has been a subject of significant research interest in cattle, particularly in Brahman cattle. A study highlighted the presence of a large-effect mutation in the PLAG1 that influences insulin-IGF concentration in Australian Brahman cattle (Bolormaa et al., 2013). This finding underscores the role of PLAG1 in regulating physiological processes related to growth and development in cattle. According to Takasuga (2015), the height-increasing allele of PLAG1 in Brahman cattle most likely originated from Bos taurus cattle via introgression and subsequent selection. The presence of Bos taurus haplotypes near the PLAG1 region on Brahman chromosomes adds to the genetic influence of PLAG1 in Brahman cattle (Fortes et al., 2013). Research on the PLAG1 in cattle, including Brahman cattle, emphasizes its role in regulating economically significant characteristics, i.e., body weight and measurements. The genetic variations and associations discovered in this study have important implications for breeding programs and genetic selection strategies in cattle, particularly in Brahman populations. ConclusionIn conclusion, g.48308C>T SNP and g.32212 19 bp indel of the PLAG1 were polymorphic. They were significantly associated with body weight and chest girth in Brahman cattle. These findings could be beneficial for the selection superior Brahman cattle in the future using marker assisted selection. AcknowledgmentsThis study was supported by the Indonesian Ministry of Research Technology and Higher Education, through the National Competitive Basic Research scheme with contract No. 221.1/UN27.22/HK.07.00/2021 and RKAT PTNBH Universitas Sebelas Maret fiscal year 2024 through Penelitian Hibah Grup Riset (Penelitian HGR-UNS) A research scheme with agreement decree No. 194.2/UN27.22/PT.01.03/2024. Also, the authors would like to convey gratitude to all staffs of the the BPTU-HPT Sembawa for the great support during this study. Conflict of interestThe authors declare that there is no conflict of interest with any financial, personal, or other relationships with other people or organization related to the material discussed in the manuscript. Author’s contributionsAll authors had participated in samples collection, performing tests, analyzing the data, and preparing and revising the manuscript. Data availabilityAll data supporting the findings of this study are available within the manuscript ReferencesAkmalputra, M.R., Sukaryo, S., Yanti, Y., Riyanto, J., Bramastya, T.A., Volkandari, S.D., Sudrajad, P. and Cahyadi, M. 2022. Identification of the PLAG1 polymorphisms in Peranakan Ongole cattle population. IOP Conf. Ser. Earth Environ. Sci. 1115, 012002. Allendorf, F.W. and Luikart, G.H. 2007. Conservation and the genetics of population. Oxford, UK: Blackwell Publishing. Bilyaro, W., Rafian, T. and Lase, J.A. 2023. Application of genetics in animal production improvement efforts in an effort to increase food production of animal origin. J. Agric. Anim. Sci. 3(2), 70–77. Bolormaa, S., Pryce, J.E., Kemper, K., Savin, K., Hayes, B.J., Barendse, W., Zhang, Y., Reich, C.M., Mason, B.A., Bunch, R.J., Harrison, B.E., Reverter, A., Herd, R.M., Tier, B., Graser, H.U. and Goddard, M.E. 2013. Accuracy of prediction of genomic breeding values for residual feed intake and carcass and meat quality traits in Bos taurus, Bos indicus, and composite beef cattle. J. Anim. Sci. 91(7), 3088–3104. Bramastya, T.A., Sukaryo, S., Dhiaurridho, M.I., Riyanto, J., Volkandari, S.D., Sudrajad, P. and Cahyadi, M. 2022. Characteristics of body weight and measurement of Peranakan Ongole and Brahman cattle in the tropics. IOP Conf. Ser. Earth Environ. Sci. 1001, 012015. Cahyadi, M., Sukaryo, S., Dhiaurridho, M.I., Bramastya, T.A., Yanti, Y., Riyanto, J., Volkandari, S.D. and Sudrajad, P. 2022. Association of pleomorphic adenoma gene 1 with body weight and measurement of Bali cattle (Bos javanicus). Vet. World. 15(3), 782−788. Cominotte, A., Fernandes, A.F.A., Dorea, J.R.R., Rosa, G.J.M., Ladeira, M.M., Van Cleef, E.H.C.B., Pereira, G.L., Baldassini, W.A. and Machado Neto, O.R. 2020. Automated computer vision system to predict body weight and average daily gain in beef cattle during growing and finishing phases. Livest. Sci. 232, 103904. Engle, B.N. and Hayes, B.J. 2022. Genetic variation in PLAG1 is associated with early fertility in Australian Brahman cattle. J. Anim. Sci. 100(4), 1−8. Fathira, A., Noor, R.R. and Jakaria, J. 2022. Diversity of SNP c.795A>G PLAG1 gene and its association to birth weight of Bali cattle. Indonesian J. Anim. Vet. Sci. 27(3), 107–113. Fortes, M.R.S., Kemper, K., Sasazaki, S., Reverter, A., Pryce, J.E., Barendse, W., Bunch, R., McCulloch, R., Harrison, B., Bolormaa, S., Zhang, Y.D., Hawken, R.J., Goddard, M.E. and Lehnert, S.A. 2013. Evidence for pleiotropism and recent selection in the PLAG1 region in Australian beef cattle. Anim. Genet. 44, 636–647. Guo, S.W. and Thomson, E.A. 1992. Performing the xact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 48, 361–372. Handayanta, E., Rahayu, E.T. and Sumiyati, M. 2016. Analisis finansial usaha peternakan pembibitan sapi potong rakyat di daerah pertanian lahan kering: Studi kasus di wilayah Kecamatan Semin, Kabupaten Gunungkidul, Daerah Istimewa Yogyakarta. Sains Peternakan 14(1), 13–20. Hartati, H., Utsunomiya, Y.T., Sonstegard, T.S., Garcia, J.F., Jakaria, J. and Muladno, M. 2015. Evidence of Bos javanicus x Bos indicus hybridization and major QTLs for birth weight in Indonesian Peranakan Ongole cattle. BMC Genet. 16, 75. Hikmawaty, H., Bellavista, B., Mahmud, T.A.B.A. and Salam, A. 2018. Korelasi bobot badan dan variabel variabel ukuran tubuh sebagai dasar seleksi calon induk sapi Bali. AGROVITAL: J. Ilmu Pertan. 3(1), 11–13. Indrayani, I. and Andri, A. 2018. Infleuence factors of beef cattle farm’s income In Sitiung, Dharmasraya Distric. J. Peternak. Indones. 20(3), 151–159. Kamprasert, N., Duijvesteijn, N. and Van der Werf, J.H.J. 2019. Estimation of genetic parameters for BW and body measurements in Brahman cattle. Animal 13(8), 1576–1582. Lachance, J. 2016. Hardy-weinberg equilibrium and random mating. Encyclopedia of evolutionary biology. 208–211. Maskur, C.A., Afikasari, D. and Ervandi, M. 2023. Critical review of beef cattle breast cattle problems In Probolinggo District. J. Sains Ternak Tropis 1(2), 54–64. Mia, L., Mony, S.I., Maruf, T.M., Pabitra, M.H., Bhuiyan, A.K.F.H., Talukder, K.U., Motaleb, M.A. and Bhuiyan, M.S.A. 2022. Detection of 19-bp indel of PLAG1 gene and its effects on morphometric traits in indigenous and crossbred cattle of Bangladesh. Bangladesh J. Anim. Sci. 51(1), 31−39. Naserkheil, M., Lee, D.H. and Mehrban, H. 2020. Improving the accuracy of genomic evaluation for linear body measurement traits using single-step genomic best linear unbiased prediction in Hanwoo beef cattle. BMC Genet. 21, 144. Nei, M. and Kumar, S. 2000. Molecular evolution and phylogenetics. Oxford, UK: Oxford University Press. Novita, R., Karyono, T. and Rasminah, R. 2019. Kualitas semen sapi Brahman pada persentase tris kuning telur yang berbeda. J. Sain Peternakan Indones. 14(4), 351–358. Pereira, A.G.T., Utsunomiya, Y.T., Milanesi, M., Torrecilha, R.B.P., Carmo, A.S., Neves, H.H.R., Carvalheiro, R., AjmoneMarsan, P., Sonstegard, T.S., Sölkner, J., Contreras-Castillo, C.J. and Garcia, J.F. 2016. Pleiotropic genes affecting carcass traits in Bos indicus (Nellore) cattle are modulators of growth. PLoS One 11, e0158165. Priyadi, D.A., Panjono, P., Bintara, S. and Hartatik, T. 2017. Genotype of Brahman and Brahman cross cattle based on SNP in insulin-like growth factor binding protein-3 (IGFBP-3) gene sequences. Biodiversitas 18(2), 795–800. Rusdiana, S. and Praharani, L. 2018. Development of people’s people livestock: SWAT private vocational policy and feasibility of animal businesses. Forum Penelitian Agro-Ekonomi 36(2), 97–116. Sholikah, N., Sutomo, A., Widiasmoro, N.P., Wahjuningsih, S., Yekti, A.P.A., Kuswati, K. and Susilawati, T. 2018. Hubungan antara tingkah laku seksual dengan produksi spermatozoa sapi Brahman. J. Agripet. 18(2), 67–73. Song, Y., Xu, L., Chen, Y., Zhang, L., Gao, H., Zhu, B., Niu, H., Zhang, W., Xia, J., Gao, X. and Li, J. 2016. Genome-wide association study reveals the PLAG1 gene for knuckle, biceps and shanks weight in Simmental Beef cattle. PLoS One 11(12), e0168316. Suhada, H., Sumadi, S. and Ngadiyono, N. 2009. Estimasi parameter genetik sifat produksi sapi Simmental di Balai Pembibitan Ternak Unggul Sapi Potong Padang Mengatas Sumatera Barat. Buletin Peternakan 33(1), 1−7. Sukaryo, S., Akmalputra, M.R., Romadhon, S., Yanti, Y., Volkandari, S.D., Sudrajad, P., Bramastya, T.A. and Cahyadi, M. 2023. Polymorphisms of the PLAG1 and their relationships with body measurments and carcass traits of Ongole crossbred cattle. IOP Conf. Ser. Earth Environ. Sci. 1183, 012014. Sukaryo, S., Augustin, R., Yanti, Y., Riyanto, J., Volkandari, S.D., Sudrajad, P. and Cahyadi, M. 2022. Identification of 19-bp indel of the Pleomorphic Adenoma Gene 1 in Bali cattle population. E3S Web Conf. 335, 00011. Takasuga, A. 2015. PLAG1 and NCAPG-LCORL in livestock. Anim. Sci. J. 87(2), 159–167. Utsunomiya, Y.T., Milanesi, M., Utsunomiya, A.T.H., Torrecilha, R.B.P., Kim, E.S., Costa, M.S., Aguiar, T.S., Schroeder, S., Carmo, A.S., Carvalheiro, R., Neves, H.H.R., Padula, R.C.M., Sussai, T.S., Zavarez, L.B., Cipriano, R.S., Caminhas, M.M.T., Hambrecht, G., Eufemi, L.C.E., Marsan, P.A., Cesana, D., Sannazaro, M., Buora, M., Morgante, M., Liu, G., Bickhart, D., Tassell, C.P.V., Sölkner, J., Sonstegard, T.S. and Garcia, J.F. 2017. A PLAG1 mutation contributed to stature recovery in modern cattle. Sci. Rep. 7(1), 1–15. Xu, W., He, H., Zheng, L., Xu, J.W., Lei, C.H., Zhang, G.M., Dang, R.H., Nui, H., Qi, X.L., Chen, H. and Huang, Y.Z. 2018. Detection of 19-bp deletion within PLAG1 gene and its effect on growth traits in cattle. Gene 675, 144−149. Zhang, Z., Yang, P., He, P., Xu, J., Lyu, S., Liu, X., Cai, C., Li, H., Li, Z., Ru, B., Xie, J., Lei, C., Chen, H., Wang, E. and Huang, Y. 2020. Distribution and association study of PLAG1 gene between copy number variation and Chinese cattle populations. Anim. Biotechnol. 33(2), 273−278. Zhong, J.L., Xua, J.W., Wanga, J., Wena, Y.F., Niub, H., Zheng, L, Hea, H., Penga, K., Hea, P., Shia, S.Y., Huanga, Y.Q., Leia, C.Z., Danga, R.H., Lana, X.Y., Qid, X.L., Chena, H. and Huanga, Y.Z. 2019. A novel SNP of PLAG1 gene and its association with growth traits in Chinese cattle. Gene 689, 166−171. Zhou, Z., Huang, B., Lai, Z., Li, S., Wu, F., Qu, K., Jia, Y., Hou, J., Liu, J., Lei, C. and Dang, R. 2019. The distribution characteristics of a 19-bp indel of the PLAG1 gene in Chinese cattle. Animals 9, 1082. | ||
| How to Cite this Article |
| Pubmed Style Trilaksono B, Rodhiyah AF, Yanti Y, Riyanto J, Kurniawan H, Imron M, Nista D, Ginto Y, Sudrajad P, Cahyadi M. Body weight and measurement traits of Brahman cattle affected by pleomorphic adenoma gene 1 (PLAG1) variations. Open Vet. J.. 2024; 14(12): 3248-3256. doi:10.5455/OVJ.2024.v14.i12.10 Web Style Trilaksono B, Rodhiyah AF, Yanti Y, Riyanto J, Kurniawan H, Imron M, Nista D, Ginto Y, Sudrajad P, Cahyadi M. Body weight and measurement traits of Brahman cattle affected by pleomorphic adenoma gene 1 (PLAG1) variations. https://www.openveterinaryjournal.com/?mno=214897 [Access: January 11, 2026]. doi:10.5455/OVJ.2024.v14.i12.10 AMA (American Medical Association) Style Trilaksono B, Rodhiyah AF, Yanti Y, Riyanto J, Kurniawan H, Imron M, Nista D, Ginto Y, Sudrajad P, Cahyadi M. Body weight and measurement traits of Brahman cattle affected by pleomorphic adenoma gene 1 (PLAG1) variations. Open Vet. J.. 2024; 14(12): 3248-3256. doi:10.5455/OVJ.2024.v14.i12.10 Vancouver/ICMJE Style Trilaksono B, Rodhiyah AF, Yanti Y, Riyanto J, Kurniawan H, Imron M, Nista D, Ginto Y, Sudrajad P, Cahyadi M. Body weight and measurement traits of Brahman cattle affected by pleomorphic adenoma gene 1 (PLAG1) variations. Open Vet. J.. (2024), [cited January 11, 2026]; 14(12): 3248-3256. doi:10.5455/OVJ.2024.v14.i12.10 Harvard Style Trilaksono, B., Rodhiyah, . A. F., Yanti, . Y., Riyanto, . J., Kurniawan, . H., Imron, . M., Nista, . D., Ginto, . Y., Sudrajad, . P. & Cahyadi, . M. (2024) Body weight and measurement traits of Brahman cattle affected by pleomorphic adenoma gene 1 (PLAG1) variations. Open Vet. J., 14 (12), 3248-3256. doi:10.5455/OVJ.2024.v14.i12.10 Turabian Style Trilaksono, Baharudin, Amira Fathin Rodhiyah, Yuli Yanti, Joko Riyanto, Hendra Kurniawan, Muhammad Imron, Delly Nista, Yumoko Ginto, Pita Sudrajad, and Muhammad Cahyadi. 2024. Body weight and measurement traits of Brahman cattle affected by pleomorphic adenoma gene 1 (PLAG1) variations. Open Veterinary Journal, 14 (12), 3248-3256. doi:10.5455/OVJ.2024.v14.i12.10 Chicago Style Trilaksono, Baharudin, Amira Fathin Rodhiyah, Yuli Yanti, Joko Riyanto, Hendra Kurniawan, Muhammad Imron, Delly Nista, Yumoko Ginto, Pita Sudrajad, and Muhammad Cahyadi. "Body weight and measurement traits of Brahman cattle affected by pleomorphic adenoma gene 1 (PLAG1) variations." Open Veterinary Journal 14 (2024), 3248-3256. doi:10.5455/OVJ.2024.v14.i12.10 MLA (The Modern Language Association) Style Trilaksono, Baharudin, Amira Fathin Rodhiyah, Yuli Yanti, Joko Riyanto, Hendra Kurniawan, Muhammad Imron, Delly Nista, Yumoko Ginto, Pita Sudrajad, and Muhammad Cahyadi. "Body weight and measurement traits of Brahman cattle affected by pleomorphic adenoma gene 1 (PLAG1) variations." Open Veterinary Journal 14.12 (2024), 3248-3256. Print. doi:10.5455/OVJ.2024.v14.i12.10 APA (American Psychological Association) Style Trilaksono, B., Rodhiyah, . A. F., Yanti, . Y., Riyanto, . J., Kurniawan, . H., Imron, . M., Nista, . D., Ginto, . Y., Sudrajad, . P. & Cahyadi, . M. (2024) Body weight and measurement traits of Brahman cattle affected by pleomorphic adenoma gene 1 (PLAG1) variations. Open Veterinary Journal, 14 (12), 3248-3256. doi:10.5455/OVJ.2024.v14.i12.10 |