| Research Article | ||
Open Veterinary Journal, (2024), Vol. 14(1): 200-213 Original Research Transcriptional responses of cytokines, immunoglobulin A, and nitric oxide genes in 1-day-old chicks post Salmonella typhimurium infection: An experimental studyYasmine I. Mosa1†*, Ahlam A. Gharib†1, Sara Y. Abd-El Galil1, Amira. M. Ali2 and Etab M. Abo Remela3,41Microbiology Department, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Sharkia, Egypt 2The Veterinary Hospital, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Sharkia, Egypt 3Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Kafrelsheikh University, Kafrelsheikh, Egypt 4Department of Biology, College of Science, Taibah University, Medina, Saudi Arabia †These authors contributed equally to this work *Corresponding Author: Yasmine I. Mosa. Microbiology Department, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Sharkia, Egypt. Email: yi.mohamed23 [at] vet.zu.edu.eg Submitted: 01/10/2023 Accepted: 15/12/2023 Published: 31/01/2024 © 2024 Open Veterinary Journal
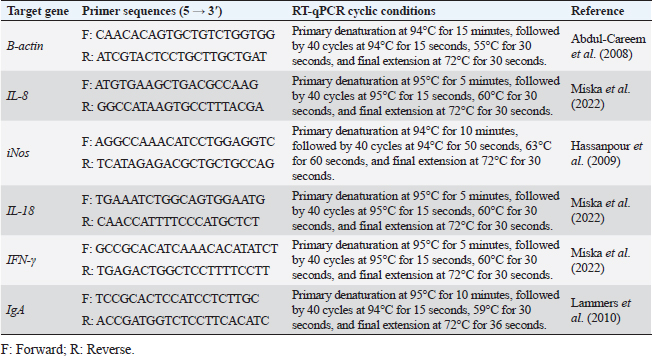
AbstractBackground: Salmonella has become one of the hazards prevalent foodborne pathogens causing different diseases in chickens. However, Salmonella typhimurium (ST), a nonhost-specific serovar, is a major avian agent that causes severe disturbance in young chicken wellness. Aim: The occurrence of Salmonella in chickens and their antimicrobial resistance were explored in this study. In addition, the immune response of 1-day-old broiler chicks, against multidrug resistant (MDR) ST infection, was also assessed at 4 and 24 hours post infection (pi) in the cecum and spleen, representing their mucosal and systemic immune responses, respectively. Methods: A total of 375 samples from 130 diseased and apparently healthy broiler and layer chickens were randomly collected for Salmonella isolation, identification, and resistance profile evaluation, from farms and different clinical laboratories. The immune response of 1-day-old broiler chicks, Ross 308, against in-vivo ST infection was ascertained through the evaluation of heterophile phagocytosis and s expression of cytokines, immunoglobulin A and other immune-regulating genes in the cecum and spleen. Twenty-four, 1-day-old nonvaccinated broiler chicks were used and divided into two groups. The chicks in the infected group were orally inoculated with 0.5 ml of 2 × 108 colony forming units (CFU)/ml of MDR ST suspension, while those in the control group were taken nutrient broth. Results: Seven out of 130 (5.38%) examined chickens were positive for Salmonella. All isolates (100%) were resistant to amoxicillin–clavulanic acid (AMC), cefazolin (CZ), cefoxitin (FOX), ciprofloxacin (CIP), nalidixic acid (NA), tetracycline (TE), fosfomycin (FOS), and colistin (CT) with multiple antimicrobial resistances (MARs) index range of 0.72–0.83, where none of them was resistant to meropenem (MEM). The results of immune response revealed that chicks infected with ST showed significantly different phagocytosis percentages and index values compared to controls. According to the real-time quantitative polymerase chain reaction (RT-qPCR) results, the transcription of IL-8, iNOS, IL-18, IgA, and IFN-γ for chicks infected by ST showed a significantly increased trend (p < 0.01) with increasing chicken age and was higher in the cecum than spleen compared to controls (p < 0.05) during 24 hours after infection. Conclusion: The findings indicated a strong mucosal immune response in the chicks after the ST challenge, which reflects humoral and cellular responses. Our insight recommended the occurrence of a natural immune response stimulator at 1 day age to face the infection, and this can prevent the resistance transfer, with efficient control measures. Keywords: Salmonella typhimurium, Cytokines, Challenge, Phagocytosis, Real-time PCR. IntroductionSalmonella is a common microorganism that can live in the intestines of animals, including birds. It can be passed to humans through contaminated water or food causing a variety of illnesses, from mild diarrhea to severe food poisoning. Salmonella infections are a serious problem for community health, animal welfare, and food safety around the world (Jajere, 2019; Hassan et al., 2021). Salmonellosis is a broad term that encompasses all infections caused by Salmonella bacteria. These infections can range from mild food poisoning to more serious invasive infections. Invasive infections can systematically reach many host parts, such as the bloodstream. Food poisoning caused by Salmonella is usually less serious, but it can still cause symptoms such as diarrhea, vomiting, and fever (Fàbrega and Vila, 2013). There are over 2,500 different types of Salmonella bacteria, and about 2,000 of them have been found in chickens. This means that chickens are the main cause of human salmonellosis (Ramatla et al., 2020). Avian salmonellosis is an infectious disease that has two forms: acute and chronic. Many types of birds may be infected by highly specialized serotypes such as Salmonella Gallinarum, S. Pullorum, and S. Arizona, or by zoonotic species such as Salmonella typhimurium (ST), Salmonella enterica (SE), and further paratyphoid Salmonella serotypes (Abd EL-Ghany et al., 2012; Wibisono et al., 2020). A genetic relationship was found between Salmonella of chicken origin and that of infected humans with Salmonella food poisoning. Chickens receive Salmonella serotypes infection either by cross-contamination with feces, water, tools, and workers or from infected birds (Meteab and Abed, 2018; Shen et al., 2023). The chicken industry has been found to be responsible for up to 50% of salmonellosis outbreaks (Antunes et al., 2016). Infection with SE in chickens can happen at every age, although it is most common in young chicks because they are more susceptible to infection (Berndt et al., 2007). Adult birds are more resilient to salmonellosis than young age because they have stronger immune systems that are able to fight off the infection and with well-developed humoral and cell-mediated immune responses (Beal et al., 2004). The serovar of ST can cause serious illness in both chickens and humans. In chickens, it can cause severe intestinal inflammation and diarrhea, leading to death in young birds. In humans, it can cause acute gastrointestinal disease, with symptoms such as diarrhea, vomiting, and fever. This bacterial agent is the most important challenge for the poultry economy, as it can lead to the loss of chickens and the contamination of poultry products (Dar et al., 2019). The intestinal mucosal immune system is the host’s major line of defense against Salmonella infection. This system includes antibodies called IgA, as well as lymphocytes and leukocytes that are found in the lining of the intestine. These cells work together to prevent Salmonella from entering the body and causing infection (Sheela et al., 2003). Phagocytosis is an innate immune response that is primarily a response to body infection (Sornplang et al., 2015). It is a process by which the cells engulf and destroy foreign particles, including pathogens and dead cells. It is a vital immune response part that plays a significant role in maintaining host health. Phagocytes, such as neutrophils and macrophages, are able to recognize and engulf foreign particles through a process called receptor-mediated endocytosis (Rosales and Uribe, 2017). Chicken heterophils, a type of white blood cell, are able to engulf and destroy all types of pathogens. After a pathogen is engulfed, the heterophil releases granules that contain enzymes and other chemicals. These chemicals kill the pathogen and also produce an oxidative burst, which is a release of free radicals that can also damage the pathogen (Genovese et al., 2013). After Salmonella bacteria invaded the gut mucosa and localized there, immune cells were detected in the cecum by immunohistochemistry from 4 hours to 9 days post infection (pi). The number of TCR1+ cells, granulocytes, CD8+ cells, and the gene expression fold change for IL-18, TNF-α, IL-12, and iNOS in the cecum increased with the tissue invasiveness of Salmonella (Berndt et al., 2007). Cytokines are proteins that are released by the immune cells and show a vital role in the immune system. They are the envoys that initiate as well as manipulate immune responses, helping to destroy microbial infections. Cytokines are also involved in inflammation, which is the body’s response to injury or infection (Charles et al., 2001). Chemokines are a kind of cytokine that has chemoattractant properties and regulates the migration of immune cells ( Kaiser and Staheli, 2008; Kaiser et al., 2022). In chickens, the gut immune response begins to interact with the commensal microbiota shortly after hatching. This interaction results in inflammation with low levels, which is related to up-regulation of the cytokine IL-8 gene expression. IL-8 is a protein that helps to recruit immune cells to the gut, which helps to protect the chicken from infection (Bar-Shira and Friedman, 2006). In chickens, IL18 is a protein that is released by intestinal epithelial cells. It induces a vital share in defending the chicken against infection by attracting immune cells to the gut, helping to kill pathogens, and stimulating the adaptive immune response. IL18 and iNOS are also released by macrophages, where they help to kill pathogens by creating nitric oxide and reactive oxygen species. IFNγ is a protein that is released from γδ T cells and natural killer (NK) cells. It helps to destroy the infected cells, stimulate the immune response, and attract other immune cells to the infection site (Ijaz et al., 2021). Understanding the mechanisms of Salmonella infection, intestinal colonization, persistence, and excretion in feces was previously discussed and proved critical (Andino and Hanning, 2015). The evaluation of chicken mucosal or systemic immune response, at 1 day old, is important to face Salmonella infection in poultry. This knowledge is essential for developing effective measures to exaggerate immune response and/or to decrease Salmonella infection in poultry farms and then salmonellosis risk in humans. Herein, we investigated the occurrence of Salmonella in Egyptian chickens and their antimicrobial susceptibilities to various antimicrobial agents. Furthermore, the immune response of 1-day-old broiler chicks was assessed against in-vivo ST infection by evaluation of heterophil phagocytosis and mRNA expression of certain cytokines, IgA, and other immune regulating genes in both cecum and spleen. Materials and MethodsSamplingThis study was conducted during the period from April to September 2022. A total of 375 chicken organs including 130 livers, 130 cecum, and 115 spleens of 130 diseased and apparently healthy broiler (n=75) and layer (n=55) chickens were included in the study. The chickens (Ages ranged from 1 to 30 days) were randomly collected from chicken farms and different clinical laboratories (private laboratories and Animal Health Research Institute, Zagazig branch) in Sharkia Province, Egypt. The samples were put in sterile vessels and transferred as early as possible to the Bacteriological Laboratory, Microbiology Department, Faculty of Veterinary Medicine, Zagazig University, where they were subjected to routine bacteriological examination for Salmonella isolation. The investigation was recommended and approved by the Zagazig University Institutional Animal Care and Use Committee (ZU-IACUC/2/F/76/2022). Salmonella isolation and identificationSalmonella isolation from the samples was carried out based on the protocol of ISO, the International Organization for Standardization (ISO, 2017). First, one gram of each organ sample was suspended in 9 ml of buffered peptone water (BPW) (Oxoid, UK) and incubated at 37 °C for 18 ± ٢ hours. After the pre-enrichment, 0.1 ml of culture was inoculated into 10 ml broth of Rappaport Vassiliadis (RV) (Oxoid, United Kingdom) and incubated at 42°C for 24 hours. The plates were cultivated on xylose lysine deoxycholate agar (XLD) (Oxoid, UK) and salmonella shigella agar (SS) (Oxoid, UK) then they were incubated at 37°C/24-hours. For the purpose of biotyping, colonies from each fresh pure culture were examined for urease production and for their growth on triple sugar iron (Oxoid, United Kingdom) media. All isolates were serotyped using commercial antisera (Denka Seiken Company Ltd., UK) (Kauffmann, 1957) via slide agglutination tests at the serology unit, Animal Health Research Institute, Dokki, Giza. Molecular identification of Salmonella isolates was confirmed by polymerase chain reaction (PCR) using the genus (invA) and species (ST fliC)—specific primers at the Biotechnology unit, Animal Health Research Institute, Zagazig branch. The amplification for invA gene was conducted using the primer set of S139-F: 5′-GTGAAATTATCGCCACGTTCGGGCAA-3′and S141-R: 5′-TCATCGCACCGTCAAAGGAACC-3′, with thermocycling conditions consisting of an early step at 94°C for 5 minutes followed by 35 cycles at 94°C for 30 seconds, 55°C for 30 seconds, 72°C for 30 seconds, then a last extension step at 72°C for 7 minutes (Oliveira et al., 2003). The amplification for ST fliC gene was conducted using the primer pair of Fli15-F: 5′-CGGTGTTGCCCAGGTTGGTAAT-3′ and Tym-R: 5′-ACTGGTAAAGATGGCT-3′, with thermocycling conditions consisting of an early step at 94°C for 3 minutes, followed by 30 cycles of at 94°C for 1 minutes, 55°C for 1 minutes, 72°C for 1.5 minutes, then a last extension step at 72°C for 10 minutes (Halimi et al., 2014). SE subspecies enterica serovar Typhimurium derived from ATCC® 14028 was used for quality control. Antimicrobial sensitivity testing of Salmonella isolatesThe recovered Salmonella isolates were examined for their susceptibilities against 18 commonly used antimicrobial agents (representing ten antimicrobial groups) by the disc diffusion method using Mueller–Hinton agar (Oxoid, UK) media. The examined antimicrobial agents including: ampicillin (AMP, 10 µg), amoxycillin-clavulanic acid (AMC, 20/10 µg), cefazolin (CZ, 30 µg), cefepime (FEP, 30 µg), ceftriaxone (CRO, 30 µg), cefoxitin (FOX, 30 µg), imipenem (IPM, 10 µg), meropenem (MEM, 10 µg), tobramycin (TOB, 10 µg), gentamicin (CN, 10 µg), amikacin (AK, 30 µg), ciprofloxacin (CIP, 5 µg), nalidixic acid (NA, 30 µg), tetracycline (TE, 30 µg), fosfomycin (FOS, 50 µg), chloramphenicol (C, 30 µg), sulfamethoxazole-trimethoprim (SXT, 23.75/1.25 µg), and colistin (CT, 10 µg). The minimum inhibitory concentration (MIC) for CT was determined by the broth microdilution assay (Rankin, 2005). The inhibition zone size around the antimicrobial discs and interpretation for MICs were deduced based on the guidelines of CLSI, the Clinical and Laboratory Standards Institute (CLSI, 2020). Multidrug-resistant (MDR) Salmonella isolates were ascertained when the isolates were resistant to ≥3 different antimicrobial classes. The multiple antimicrobial resistance (MAR) index of each antimicrobial was calculated by the following formula: Number of resistance recorded/(Total number of examined antimicrobials × Total number of ST isolates), whereas MAR index for each ST isolate=Number of antimicrobials to which the isolate showed resistance/Number of antimicrobials to which the isolate had been examined (Tambekar et al., 2006). ST ATCC® 14028 was used for quality control. ST challenge in 1-day-old chicksChicks and experimental design Genetically distant chicks originated from the specific-pathogen-free Ross 308 line were used. Totally, 24 broiler chicks of 1 day age received no vaccination and tested to be free from Salmonella, were separated into two groups (infected and control), and kept under intense hygienic circumstances in the experimental unit of the Faculty of Veterina ry Medicine, Zagazig University. The chicks in the infected group were separately and orally challenged with 0.5 ml of 2 × 108 CFU/ml ST suspension through crop instillation by a syringe, while the control group chicks were neither immunized nor infected as shown in a previous protocol (Berndt et al., 2007) with some modifications. Bacterial isolate for the challenge One pathogenic MDR ST isolate recovered during the study was used for the challenge. It was identified phenotypically then serotyped and confirmed by PCR. Before the experimental trial, it was subjected to virulence exaltation via chicken inoculation (Berndt et al., 2007) with some modifications. The bacterial suspension for inoculation was prepared from a fresh colony inoculated into the nutrient broth (Oxoid, UK) and then incubated with shaking for 18 hours/37°C. Sample collection after challenge After two-timing intervals of 4 and 24 hours pi, blood was sampled from the wing veins of each chick (inoculated and controlled) for phagocytosis activity according to the method described previously ( Lowry et al., 2005). Moreover, tissue samples (one cecum and spleen) were collected from each euthanized bird (Berndt et al., 2007). The samples were stored in RNA-later until they were used for real-time quantitative PCR (RT-qPCR). Measurement of phagocytic activityPreparation of Candida glabrata (C. glabrata) Heat-inactivated C. glabrata (obtained from the Microbiology Department, Mycology unit, Faculty of Veterinary Medicine, Zagazig University) was used to evaluate the phagocytic activity of chick’s heterophils. Candida glabrata was cultivated on Sabouraud’s dextrose agar (Oxoid, UK) for 24 hours at 37°C. The grown yeast cells were harvested in sterile PBS, washed, and pelleted at 1,500 rpm for 10 minutes. The pellet was washed and adjusted to a concentration of 4 X 106 CFU / ml The yeast cells were inactivated in a water bath at 100 °C/30 minutes (Sheena and Arthur, 1978). Separation and preparation of chick’s leukocytes (heterophil isolation) Heparinized blood samples were used for leukocyte separation according to the method described by Lowry et al. (2005) with some modifications. Collected blood samples were diluted 1:2 in sterile heparinized PBS under sterile procedures, and the diluted blood was carefully layered over an equal volume of Ficoll–Hupaque solution (1.077 density gradient) (Sigma, USA) in sterile plastic centrifuge tubes. The over-layered blood was pelleted at 1,000 rpm/30 minutes. Thereafter, leukocytes in the white opaque band were aspirated using a sterile Pasteur pipette at the Histopaque plasma interface and placed in sterile Hank’s Balanced Salt Solution (HBSS) (Wadsworth Center Media Core). The separated cells were washed three times in sterile HBSS, by centrifugation for 10 minutes at 2,500, 1,500 rpm then 1,000 rpm, separately. The pelleted cells were put in 2 ml media of RPMI 1640 (Oxoid, UK) with 5% chick serum and then counted using a hemocytometer (Weber Scientific comp., Hamilton). The cells were examined for viability and their concentration was adjusted to 1 × 106cell/ml. Estimation of phagocytic percentage and phagocytic index (PI) in the experimental groups The phagocytosis activity was done as described previously (Sornplang et al., 2015; Lowry et al., 2005), with modifications. An equal amount of leukocyte suspension along with heat-inactivated C. glabrata was mixed and then incubated at 27°C/30 minutes in a humidified 5% carbon dioxide incubator. After incubation, they were centrifuged at 2,500 rpm/5 minutes. A small amount of the sediment was re-suspended and smeared onto a slide. The slide was then stained and viewed under an ordinary microscope. Heterophils were counted in ten microscopic fields, and the yeast cells were also recorded in each phagocytic cell. Phagocytosis and PI values were calculated from 200 heterophils in different fields. The phagocytic activity was assessed based on these equations (Lowry et al., 2005): P%=Number of phagocytes with engulfed yeast cells/Total number of phagocytes × 100, PI=Total number of engulfed yeast cells/Total number of phagocytes with engulfed yeast. Tissue cytokines and other immune gene expressions by RT-qPCR The extraction of RNA was carried out using the QIAamp RNeasy Mini kit (Qiagen, Germany, GmbH) according to the manufacturer’s instructions. For SYBR green RT-qPCR, the specificity of amplified products was confirmed by generating melting curves, primer sets (Metabion, Germany) used for the amplification of IFNγ, IL-8, IL-18, iNOS, IgA, and β-Actin genes and their cyclic conditions were described in Table 1. The relative expression was performed in MX3005P real-time PCR thermal cycler (Agilent, La Jolla, CA, United States) using QuantiTect SYBR Green real-time PCR Kit (Qiagen, Germany) according to the manufacturer’s recommendations. The β-Actin gene was used as a normalizer. The CT value of each sample was then compared to the value of the positive control using the method previously described (Yuan et al., 2006) and the fold change values were estimated using the 2-▫▫ct method. Statistical analysisMicrosoft Office Excel (Microsoft Corporation, Redmond, WA) was used for data amending. Normality and homogeneity of variance were conducted by the Levene and Shapiro–Wilk tests (Razali and Wah, 2011). The significant effects of treatment, time, and their interaction on chicken phagocytic activity after Salmonella challenge were examined by conducting A two-way ANOVA (Proc ANOVA) (SAS, 2012). The mixed model of the statistical analysis system (SAS, 2012) (version 8, Cary, NC) was used for assessing gene transcription of IL-8, iNOS, IL-18, IgA, and IFN-γ for chickens infected by Salmonella spp. The statistical model includes the effect of treated time, organs, and their interactions as fixed factors, while chicks’ individuality is a random factor. Duncan’s Multiple Range Test (DMRT) (Steel and Torrie, 1980) was used for the detection of the multiple comparisons among means. Resulted data were expressed as means ± SE. Statistical significance was accepted at a probability <0.05 (p ≤0.05). Table 1. Primer sequences, target genes, and cycling conditions for SYBR green Rt-qPCR.
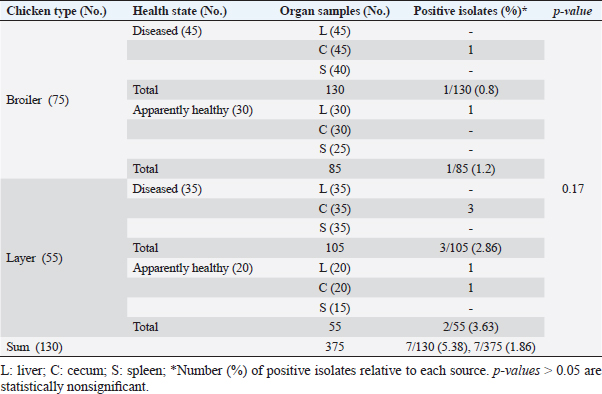
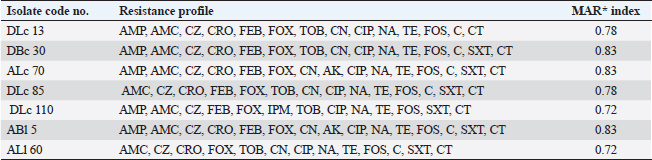
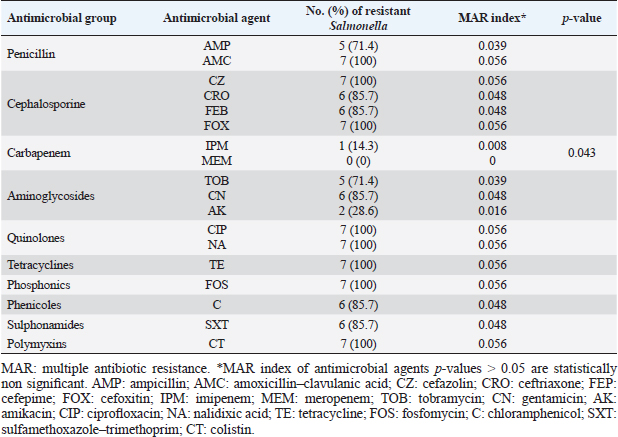
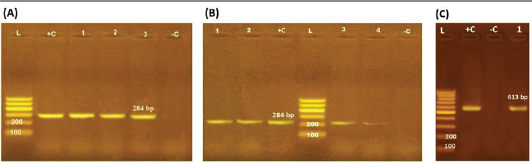
ResultsOccurrence of Salmonella in chicken samplesSeven positive Salmonella isolates were recovered from 130 chickens (7/130, 5.38% as 7/375, 1.86% of total samples); four of them were from diseased chicken samples, and three were from apparent healthy ones, mostly they were recovered from the cecum (Table 2). Traditional phenotypic methods identified the isolates as Salmonella based on their characteristic appearance on XLD and SS agar media. On XLD agar, the isolates appeared as pink colonies usually with black centers, but on SS agar, they gave white colonies with black centers. Biochemically, the isolates were negative for the urease test and produced a yellow butt and red slant with black coloration on TSI agar. The isolates were consistently serotyped as Salmonella, three of them were recorded as ST (2.3%; 3/130) showing 1, 4, 5, 12:i, formula, and the other four recovered isolates showing 1, 9, 12:-:- formula (suspected to be Salmonella pullorum in young chicks). ST isolates were further genotypically confirmed at the genus level (284 bp for the invA gene), as well as at the species level (613 bp fliC gene) to be used for the experimental design (Suppl. Fig. 1). The antimicrobial sensitivity of Salmonella isolatesThe susceptibility result of seven recovered Salmonella isolates to 18 antimicrobial drugs presented that they were resistant to amoxicillin–clavulanic acid, CZ, FOX, CIP, NA, TE, FOS, and CT (100%), where none of them was resistant to MEM. A high resistance rate was observed to CRO, FEP, CN, C, and sulfamethoxazole–trimethoprim (85.7%, each), while 71.4% of the isolates were resistant to AMP and TOB. A low resistance rate was observed for AK and IPM with percentages of 28.6% and 14.3%, respectively (Table 3). The broth microdilution results indicated that the MIC value for CT ranged from 32 to 128 µg/ml. Salmonella isolates showed different MAR patterns and the MAR index of Salmonella isolates (n=7) ranged from 0.72 to 0.83 (Table 3) with an average of 0.78, while that of each antimicrobial agent ranged from 0.008 to 0.05. There is a significant difference (p < 0.05) in the MAR index between the different antimicrobial agents (Table 4). ST challenge:Salmonella challenge agentOne pathogenic MDR ST isolates (code no. DBc30) was selected for the challenge. It was serotyped by the formula: 1, 4, 5, 12: i, and molecularly confirmed. It showed antimicrobial resistance to 15 antimicrobial agents with a MAR index of 0.83 (Table 3). Clinical signs and postmortem lesions in the experimental groupsAfter ST inoculation, the challenged chicks showed noticeable clinical symptoms such as weakness, depression, dullness, ruffled feathers, anorexia, whitish diarrhea, Salmonella arch, and chicks collected together at the room corner. While the postmortem lesions included hepatomegaly with hemorrhage and congestion in the livers. The spleen showed splenomegaly in addition to hemorrhage, and congestion. The cecum was enlarged with some congestion, hemorrhage, and inflammation. Table 2. Salmonella-positive isolates from chicken samples.
Table 3. Antibiogram of Salmonella isolates (n=7) recovered chicken organs.
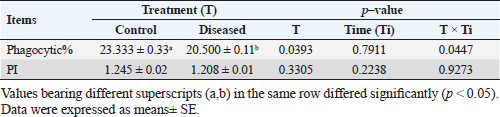
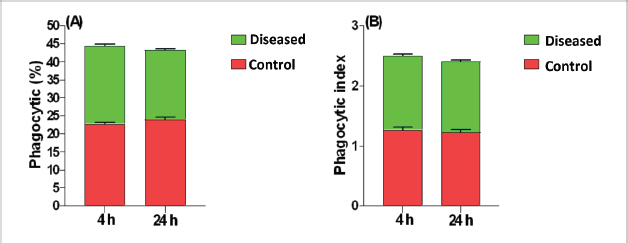
Phagocytic activity of chicks after the ST challengeThe chicks group infected with ST showed significantly different phagocytosis and phagocytosis index values at two-time intervals of 4 and 24 hours pi matched with the unchallenged control chicks. The present outcomes clearly indicated that there was a significant reduction in phagocytic activity in the challenged group compared to the control one (p=0.0393; Table 5). Regardless of the group effect, analysis of group-by-time interaction revealed that there were significant changes between the two considered groups (group of 4 hours pi and group of 24 hours pi) over time, maximizing in the control group after 24 hours pi, while the lowest phagocytosis percentage value was recorded in the diseased group during the same aforementioned time period (p < 0.05; Fig. 1). On contrary, there were no significant effects of treatment, time, or their interaction on the PI (p=0.3305, 0.2238, and 0.9273, respectively; Table 5 and Suppl. Fig. 2). This result may be due to the better or instinctive immune response in unchallenged, unacclimatized young chicks than in challenged ones. Table 4. Frequency of antimicrobial resistance in salmonella isolates (n=7) recovered from chicken organs.
Table 5. Phagocytic activity of chicks after the Salmonella challenge.
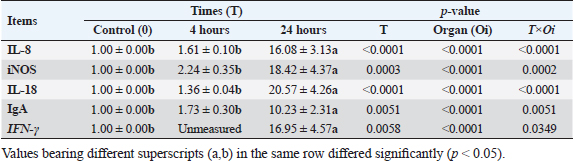
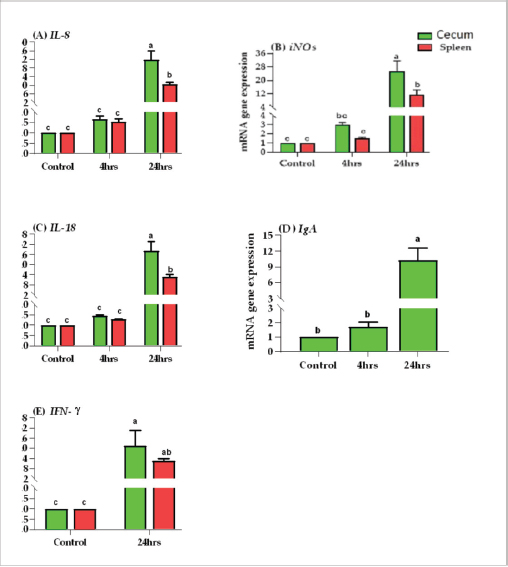
Results of tissue-specific mRNA gene expression by RT-qPCRThere were variations in the expression levels of all examined genes in infected chicks at two-time intervals compared to controls (Fig. 2). After 4 hours pi, the mRNA expression levels of IL8, IL18, iNOs, and IgA genes were higher in the cecum than the spleen; the highest mRNA expression levels in the cecum were noted for iNOS, followed by IgA, then IL8 and IL18 with fold changes of 2.9, 1.7, 1.6, and 1.4, respectively. While in the spleen, the mRNA peak expression level was recorded for IL8 followed by iNOS, then IL18, with fold changes of 1.54, 1.5, and 1.2, respectively. After 24 hours post inoculation, similarly, the mRNA expression ranks of all aforementioned genes in addition to INF-ϒ were higher in the cecum than the spleen. The mRNA peak expression levels were presented for IL18, followed by iNOS, IL8, INF-ϒ, and IgA in the cecum with fold changes of 28.2, 25.4, 21.7, 21.5, and 10.2, respectively. While in the spleen, the peak mRNA expression showed IL18 followed by INF-ϒ, iNOs, and IL8, with fold changes of 12.8, 12.3, 11.4, and 10.4, respectively. The transcription of IL-8, iNOS, IL-18, IgA, and IFN-γ for chicks infected by ST showed a significantly increased trend (p < 0.01; Table 6) with increasing chicken age. Regardless of the effect of chicken age, the analysis of treatment by organ interactions revealed that the expressions of all aforementioned genes except IgA were significantly greater in the cecum than spleen (p < 0.05) during 24 hours after infection, whereas they did not alter significantly during the 4 hours (p > 0.05; Fig. 2A–E). With respect to IgA gene expression, it was significantly up-regulated just in the cecum 24 pi compared to the control group (p < 0.05; Fig. 2E). Collectively, the present outcomes clearly indicated that the transcription of all previously detected genes (representing immune response’s stages of innate and mucosal immunities, inflammation, and Th1 of phagocytosis) was significantly higher during the 24 hours after bacterial infection than their counterparts in the control chicks (p < 0.05); however, nonsignificant changes were noticed between the control chicks and challenged ones 4 hours pi. Table 6. IL8, IL18, iNOS, IFN-ϒ. and IgA gene expression of chick organs at 4 and 24 hours post-Salmonella challenge.
Fig. 1. Treatment by time interaction affecting chicks’ phagocytic activity (A) and PI (B). DiscussionPoultry is a major reservoir of Salmonella bacteria, which can cause salmonellosis. International trade in poultry is a major factor that can facilitate the spread of Salmonella, as it can transport infected birds and their products between countries (EFSA and ECDC, 2019; Wibisono et al., 2020). In our study, seven Salmonella isolates were found in 130 diseased and apparently healthy chickens, with an occurrence percentage of 5.38%. Our results were lower than those of other studies, such as Abd El-Ghany et al. (2012), which found that Salmonella was recovered from 3.84% to 5.06% of the examined samples isolated from four chicken flocks located in El-Kalubia Governorate, Egypt; Helal et al. (2019), which found that 5.33% of the examined samples were Salmonella positive in Egypt; and Shen et al. (2023), which successfully isolated 108 (5.66%) Salmonella isolates from 1908 chicken samples in Anhui, China. However, these results were nearly similar for those of Menghistu and coauthors (Menghistu et al., 2011), who found that only 2.7% of the examined samples were Salmonella positive in India, and Medeiros team (Medeiros et al., 2011), who found that the prevalence of Salmonella in Brazil was 2.7%. However, the results of this study were lower than those of Andoh et al. (2016) (Ghana), lEl-Sharkawy et al. (2017) (Kafr El-Sheikh, Egypt), Ramatla et al. (2020) (South Africa), Ammar et al. (2016) (Sharkia Province, Egypt), and Abd El-Aziz et al. (2021) (Egypt), with recovery rates of 47%, 41%, 35%, 17%, and 11.54%, respectively. Interestingly, in the current study, all the Salmonella isolates (n=7) were 100% resistant to amoxicillin–clavulanic acid, CZ, FOX, NA, CIP, TE, FOS, and CT where none of them was resistant to MEM. A high resistance rate (85.7%) was observed to CRO, FEP, CN, C, and sulfamethoxazole–trimethoprim while resistance % to AMP and TOB was 71.4%. A low resistance rate was also observed for AK and IPM (28.6% and 14.3%, respectively). A similar resistance pattern against Salmonella isolated from different sources was observed previously (Abd El-Aziz et al., 2021). The detected highest MAR index ranged from 0.72–0.83 with an average of 0.78 was similar to that of Mir et al. (2022) who recorded that it ranged from 0.45 to 0.81 with an average of 0.63 in four Salmonella isolates that tested against 11 antimicrobial agents. The elevated MAR indices of each isolate and each antimicrobial agent pointed to the haphazard use of antibiotics.
Fig. 2. Effects of time by organ interaction (means ± SEM) on fold change of (A) IL-8, (B) iNOS, (C) IL-18, (D) IgA, and (E) IFN-γ. for Salmonella infected chicks. In this study, the phagocytosis index and phagocytosis percentage of chicks challenged with ST were lower than those of control chicks after 4 and 24 hours pi. The significantly dropped heterophil numbers might be due to stress-induced mechanisms or might be due to the death of intracellular bacteria and heterophils after Salmonella stimulation (Pieper et al., 2017). This result agreed with previous research (Sornplang et al., 2015), in which the chicks infected with SE showed lower phagocytic activity than controlled chicks at 24 and 48 hours, and another research (Sreekantapuram et al., 2021), in which the heterophils’ count in the blood of white Leghorn chickens decreased significantly after being infected with Salmonella. The number of heterophils was considerably lower (p ≤ 0.05) between 60 and 180 minutes in blood samples that were infected with SE and between 90 and 150 minutes in blood samples that were infected with Salmonella gallinarium. Salmonella bacteria initially colonize the digestive tract, especially the cecum, after being ingested orally. This can happen as early as 3 hours after infection. However, it takes about 24 hours for the bacteria to colonize the internal organs through macrophage migration (Van-Immerseel et al., 2002).
Supplementary Figure S1. Agarose gel electrophoresis pattern of PCR amplified products of Salmonella isolates genes. M: DNA molecular size marker (100 bp), +c: control positive and ˗c: control negative (A) and (B): showing PCR product of invA gene of seven isolates {three isolates of Lanes 1–3 (A) and four isolates of Lanes 1–4 (B)}; (C): showing PCR product of fliC gene of one isolate confirmed S. typhimurium (Lanes 1 is positive).
Supplementary Figure S2. Phagocytic activity of chicks after Salmonella typhimurium challenge. An arrow indicated engulfed yeast. In this research, the local and systemic immune response was evaluated after ST challenge of 1-day-old Egyptian line broiler chicks, the mRNA expression levels of IL8, IL18, iNOS, INF-ϒ, and IgA genes were measured after 4 and 24 hours pi and showed significant up-regulation at 24 hours and was higher in cecum than spleen of challenged chicken compared with control unchallenged one. The mRNA of IgA was up-regulated only in the cecum at two time points, and it is not determined in the spleen. While INF-ϒ was up-regulated only at 24 hours and data is not determined at 4 hours. Similarly, in a previous study (Withanage et al., 2004), 1-day-old chicks, after hatching, exposed to ST showed an extra rapid inflammatory cytokine response in the intestine, including the cecal tonsils, more than in the spleen. However, Beal et al. (2006) found that the immune responses at young ages after infection with Salmonella serovar Typhimurium result in a dangerous systemic response after invasion and inflammation of systemic organs. This suggests that the spleen may also play a vital role in the chicks’ immune response to Salmonella infection at young ages. A previous study (Withanage et al., 2004) demonstrated that the immune responses in systemic sites such as the liver and spleen are slower than those in the gut. This is because it takes longer for bacteria to reach these organs after they are ingested. However, chemokine expression, which is a signal that entices immune cells to the infection site, was first detected in the spleen and liver 12 hours pi, even before the bacteria had arrived. This suggests that the immune system is already preparing to fight the infection even before the bacteria have reached these organs. In the gut, the IL-8 and K60, CXC chemokines, are formed in response to infection. These chemokines attract polymorphonuclear heterophils, which are a type of white blood cells that play a role in combatting infection. The production of these chemokines and the subsequent heterophils influx into the intestine is responsible for the pathology and inflammation that is shown in the ceca and intestines. Here, the transcription of IL-8, iNOS, IL-18, IgA, and IFN- γ genes for chicks infected by ST showed a significantly increased trend (p < 0.01; Table 6) with increasing chicken ages. Between 1- and 2-days pi, the highest levels of iNOS mRNA expression were found. Furthermore, IFN-γ, the greatest significant stimulator for macrophages, increased considerably 1-day pi, with the peak of IL-18 mRNA at 2-days pi, then by the highest IFN- γ expression level at 4-days pi, and ST infected chicks produced more IFN-γ but less IL-12 and IL-18 than SE infected chicks (Berndt et al., 2007). According to the data in this study, the expression of the inflammatory genes, including IFN-γ was also elevated than in control for two weeks before returning to baseline after activating peripheral chicken blood mononuclear cells with both SE (Matulova et al., 2013) at various time points and ST (Rani et al., 2012; Dar et al., 2019). In addition, a study by Crhanova et al. (2011) found that in 1-day-old chicks infected with SE, the levels of cytokines peaked at 3 days pi but remained elevated for up to 10 days pi. IFN-γ was the only cytokine that remained up-regulated for this duration. The authors of the study proposed that the immune response of chickens infected with Salmonella shifts from a Th1 response, which is described by the creation of iNOS and IFN-γ, to a Th17 response, which is described by the creation of interleukin-17 (IL-17). The present work clearly indicated that the transcription of all previously detected genes, which represent immune response stages of innate immunity, inflammation, and Th1 of phagocytosis including; iNOS, IL8, and INF-ϒ; and IL18, respectively, were significantly higher during 24 hours after bacterial infection than their counterparts in the control group (p < 0.05); however, nonsignificant changes were noticed between 4 hours pi in relation to the control chicks and, similarly, the finding of immune response stages is shown in a previous study of SE ( Kaiser et al., 2022). A similar study (Dar et al., 2019) found that the IFN-γ, IL-12, and IL-18 mRNA expression heights were significantly increased in the liver, spleen, and cecum of infected birds matched to the control group. The highest IFN-γ mRNA expression was detected in infected birds on day 1 after infection in the liver, and up to day 5 in the spleen and cecum. On day 5 pi, the spleen had the highest increase in IFN-γ mRNA expression, followed by the cecum and liver. The author concluded that during Salmonella infection, the mRNA expression levels of different genes (IL-12, IFN-γ, and IL-18) in the spleen, liver, and cecum increase depending on the timer. Our study concentrated on the immune response first hours after the challenge in 1-day-old chick after hatching and found that the systemic immune response initiated in the spleen at 4 hours pi and increased after that till 24 hours pi; therefore, it is suggested using natural immune stimulator to face the risk of avian salmonellosis. The ascertained research result for our study is the result recorded previously (Cheeseman et al., 2007) in which the mRNA expression levels of IL-18 and interferon-gamma were higher in the spleens of chicks exposed to Salmonella. This proposes that avian splenic cells may play a role in Salmonella infection resistance. The most abundant immunoglobulin in the avian intestinal tract secretions is IgA. It protects the intestinal lining from invasion by pathogens (Tizard, 2002). In this article, the IgA mRNA expression level was measured in the cecum after 4 and 24 hours pi by ST and showed significant up-regulation. This is similar to the results of a previous study (Sheela et al., 2003), which found that oral SE inoculation of young chicks increased the number of IgA cells, the number of gut-associated T cells in the intestine, and the concentration of IgA in the intestinal mucus. On the other hand, a previous study (Beal et al., 2006) found that humoral immunity is not essential for the lasting protection or even prevention of initial avian salmonellosis. The obstacle in our study is due to limited funding resources. Therefore, the study identified the Salmonella occurrence in samples and evaluated the chick mucosal and systemic immune response at only two-time intervals using an efficient real-time PCR, but it requires various primer pairs targeting conserved genes to characterize various cytokines and other immune gene expression profiles at different age intervals after in-vivo ST infections. However, microarray is a microbiological implement used to determine the thousands of gene expressions together. ConclusionThe findings indicated a strong mucosal immune response in the chicks after the ST challenge. Chicks infected with ST showed significantly different phagocytosis percentages and phagocytosis index values compared with controls. In addition to transcription of the most examined cytokines and immune genes, mainly IL-8, iNOS, IL-18, IFN-γ, and IgA, was indicated in chick’s cecum and spleen at two-time intervals pi and was highly significant in cecum at 24 hours. Our insight recommended the occurrence of a natural immune response stimulator at 1 day age to face the infection and then prevent resistance transfer with efficient control measures. AcknowledgmentsThe authors want to thank Dr. Iman Suelam the Consultant of Microbiology at Veterinary Hospital, Zagazig University, for their great efforts in this research. Conflict of interestThe authors have no conflict of interest with the state. FundingThis article did not take any specific funding from agencies in the public or not-for-income parts. Author contributionsAG and YIM donated equally to the idea and proposal of the presented article. They also participated with SY in the submission of and use of traditional bacteriological as well as recent identification methods. YIM performed a challenging experimental design. YIM also carried out all PCR assays and participated in the analysis of their results, established the figures, and participated with AG in statistical analyses of the data. AG, YIM, SY, AA, and E.A participated in the experimental design and comprehended the study results. YIM and AG carried out the data analysis and wrote the first print of the manuscript. All authors reviewed the manuscript judgmentally for important intelligent data and gave the last agreement of the form to be available for publication. Data AvailabilityThe conclusions of this article and their data are included within the article and its supplementary files. ReferencesAbd El-Aziz, N.K., Tartor, Y.H., Gharieb, R.M.A., Erfan, A.M., Khalifa, E. and Said, M.A. 2021. Extensive drug resistant Salmonella enterica isolated from poultry and humans: prevalence and molecular determinants behind the co-resistance to ciprofloxacin and tigecycline. Front. Microbiol. 12, 738784. Abd El-Ghany, A.W., El-Shafii, A.S.S. and Hatem, M.E.A. 2012. Survey on Salmonella species isolated from chicken flocks in Egypt. Asian J. Anim. Vet. Adv. 7, 489–501. Abdul-Careem, M.F., Hunter, D.B., Thanthrige-Don, N., Haghighi, H.R., Lambourne, M.D. and Sharif, S. 2008. Cellular and cytokine responses associated with dinitrofluorobenzene-induced contact hypersensitivity in the chicken. Vet. Immunol. Immunopathol. 122, 275–284. Ammar, A.M., Mohamed, A.A., Abd El Hamid, M.I. and El-Azzouny, M.M. 2016. Virulence genotypes of clinical Salmonella Serovars from broilers in Egypt. J. Infect. Dev. Ctries. 10(4), 337–346. Andino, A. and Hanning I. 2015. Salmonella enterica: survival, colonization, and virulence differences among serovars. Sci. World J. 2015, 1–16. Andoh, L.A., Dalsgaard, A., Obiridanso, K. and Newman, M.J. 2016. Prevalence and antimicrobial resistance of Salmonella serovars Isolated from poultry in Ghana. Epidemiol. Infect. 144(15), 3288–3299. Antunes, P., Mourão, J., Campos, J. and Peixe, L. 2016. Salmonellosis: the role of poultry meat. Clin. Microbiol. Infect. 22(2), 110–121. Bar-Shira, E. and Friedman, A. 2006. Development and adaptations of innate immunity in the gastrointestinal tract of the newly hatched chick. Dev. Comp. Immunol. 30, 930–941. Beal, R.K., Powers, C., Davison, T.F., Barrow, P.A. and Smith, A.L. 2006. Clearance of enteric Salmonella enterica serovar Typhimurium in chickens is independent of B-cell function. Infect. Immun. 74, 1442–1444. Beal, R.K., Powers, C., Wigley, P., Barrow, P.A. and Smith, A.L. 2004. Temporal dynamics of the cellular, humoral and cytokine responses in chickens during primary and secondary infection with Salmonella enterica serovar Typhimurium. Avian Pathol. 33, 25–33. Berndt, A., Wilhelm, A., Jugert, C., Pieper, J., Sachse, K. and Methner, U. 2007. Chicken cecum immune response to Salmonella enterica serovars of different levels of invasiveness. Infect. Immun. 75, 5993–6007. Charles, A., Janeway, J., Travers, P., Walport, M. and Shlomchik, M.J. 2001. Immunobiology. The Immune System in Health and Disease. 5 th edition, New York, NY: Garland Science. Cheeseman, J.H., Kaiser, M.G., Ciraci, C., Kaiser, P. and Lamont, S.J. 2007. Breed effect on early cytokine mRNA expression in spleen and cecum of chickens with and without infection. Dev. Comp. Immunol. 31, 52–60. CLSI. Clinical and Laboratory Standards Institute, 2020. Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI document M100-S20. Crhanova, M., Hradecka, H., Faldynova, M., Matulova, M. and Havlickova, H. 2011. Immune response of chicken gut to natural colonization by gut microflora and to Salmonella enterica serovar enteritidis infection. Infect. Immun. 79, 2755–2763. Dar, M.A., Urwat, U., Ahmad, S.M., Ahmad, R., Kashoo, Z.A., Dar, T.A. and et al. 2019. Gene expression and antibody response in chicken against Salmonella typhimurium challenge. Poult. Sci. 98, 2008–2013. El-Sharkawy, H., Tahoun, A., El-Gohary, A.A., El-Abasy, M., El-Khayat, F., Gillespie, T. et al. 2017. Epidemiological, molecular Characterization and antibiotic resistance of Salmonella enterica serovars isolated from chicken farms in Egypt. Gut Pathogens. 9, 8. European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control (ECDC). 2019. The European Union One Health 2018 zoonoses report. EFSA J. 17(12), 5926. Fàbrega, A. and Vila, J. 2013. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 26, 308–341. Genovese, K.J., He, H., Swaggerty C.L. and Kogut M.H. 2013. The avian heterophil. Dev. Comp. Immunol. 41, 334–340. Halimi, H.A., Seifi, H.A. and Rad, M. 2014. Bovine salmonellosis in northeast of Iran: frequency, genetic fingerprinting and antimicrobial resistance patterns of Salmonella spp. Asian Pac. J. Trop. Biomed. 4, 1–7. Hassan, W.H., Hassan, H.S., Hassan, W.M., Shany, S.A. and Osman, G.S. 2021. Identification and characterization of Salmonella species isolated from broiler chickens. J. Vet. Med. Res. 28, 21–29. Hassanpour, H., Yazdani, A., Khabir Soreshjani, K. and Asgharzadeh, S. 2009. Evaluation of endothelial and inducible nitric oxide synthase genes expression in the heart of broiler chickens with experimental pulmonary hypertension. Br. Poult. Sci. 50(6), 725–732. Helal, G., Tarabees, R. and Younis, G. 2019. Molecular characterization of visrulence genes associated with Salmonella spp. Isolated from Poultry. J. Curr. Vet. Res. 2, 36–46. Ijaz, E.J.A., Veldhuizen, F., Broere, V., Rutten, C.A. and Jansen. 2021. The interplay between Salmonella and intestinal innate immune cells in chickens. Pathogens. 10, 1512. ISO. 2017. Microbiology of the Food Chain Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella. ISO 6579-1. Jajere, S.M. 2019. A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and adaptation, and antimicrobial resistance including multidrug resistance. Vet. World. 12(4), 504–521. Kaiser, M.G., Hsieh, J., Kaiser, P. and Lamont, S.J. 2022. Differential immunological response detected in mRNA expression profiles among diverse chicken lines in response to Salmonella challenge. Poult. Sci. 101, 101605. Kaiser, P. and Staheli, P. 2008. Avian cytokines and chemokines. Avian Immunol. 2008, 203–222. Kauffmann, F. 1957. The Kauffmann-White Schema; diagnostic Salmonella Antigen Schema. Copenhagen: Munksgaard 1957, 126. Lammers, A., Wieland, W.H., Kruijt, L., Jansma, A., Straetemans, T., Schots, A. and et al. 2010. Successive immunoglobulin and cytokine expression in the small intestine of juvenile chicken. Dev. Comp. Immunol. 34(12), 1254–1262. Lowry, V.K., Farnell, M.B., Ferro, P.J., Swaggerty, C.L., Bahl, A. and Kogut, M.H. 2005. Purified β-glucan as an abiotic feed additive up-regulates the innate immune response in immature chickens against Salmonella enterica serovar Enteritidis. Int. J. Food Microbiol. 98, 309–318. Matulova, M., Varmuzova, K., Sisak, F., Havlickova, H. and Babak, V. 2013. Chicken innate immune response to oral infection with Salmonella enteric serovar Eneteritidis. Vet. Res. 44, 37. Medeiros, M.A., Oliveira, D.C., Rodrigues Ddos, P. and Freitas, D.R. 2011. Prevalence and antimicrobial resistance of Salmonella in chicken carcasses at retail in 15 Brazilian cities. Rev. Panam. Salud. Publica. 30, 555–560. Menghistu, H.T., Rathore, R., Dhama, K. and Agarwal, R.K. 2011. Isolation, identification and polymerase chain reaction (PCR) detection of Salmonella Species from field materials of poultry origin. Int. J. Microbiol. Res. 2(2), 135–142. Meteab, B.K. and Abed, A.A.A. 2018. Isolation and identification of Salmonella serotypes in poultry. Al-Qadisiyah J. Vet. Med. Sci. 17, 75–80. Mir, R., Salari, S., Najimi, M. and Rashki, A. 2022. Determination of frequency, multiple antibiotic resistance index and resistotype of Salmonella spp. in chicken meat collected from southeast of Iran. Vet. Med. Sci. 8, 229–236. Miska, K.B., Kahl, S., Schreier, L.L., Russell, B., Kpodo, K.R. and Proszkowiec-Weglarz, M. 2022. Delay of feed post-hatch causes changes in expression of immune-related genes and their correlation with components of gut microbiota but does not affect protein expression. Animals 12(10), 1316. Oliveira, S.D., Rodenbusch, C.R., Cé, M.C., Rocha, S.L.S. and Canal, C.W. 2003. Evaluation of selective and non-selective enrichment PCR procedures for Salmonella detection. Lett. Appl. Microbiol. 36, 217–221. Pieper, J., Locke, M., Ruzaike, G., Voigt, S., Methner, U. and Berndt, A. 2017. In vitro and in vivo generation of heterophil extracellular traps after Salmonella exposure. Vet. Immunol. Immunopathol. 188, 1–11. Ramatla, T.A., Mphuthi, N. and Ramaili, T. 2020. Molecular detection of virulence genes in Salmonella spp. isolated from chicken faeces in Mafikeng, South Africa. J. S. Afr. Vet. Assoc. 91, 1994. Rani, S., Preeti, J., Pandey, N.K., Saxena, V.K., Saxena, M. and Singh, K.B. 2012. Cytokines expression and nitric oxide production under induced infection to Salmonella typhimurium in chicken lines divergently selected for cutaneous hypersensitivity. Asian Australas. J. Anim. Sci. 25, 1038–1044. Rankin, D.I. 2005. Test methods: MIC testing. Ed. Coyle B.M. (1st Ed.): manual of antimicrobial susceptibility testing. American Society for Microbiology, pp: 53–62. Razali, N.M. and Wah, Y.B. 2011. Power comparisons of shapiro-wilk, kolmogorov-smirnov, lilliefors and anderson-darling tests. J. Stat. Modeling Anal. 2, 21–33. Rosales, C. and Uribe-Querol, E. 2017. Phagocytosis: a fundamental process in immunity. BioMed Res. Int. 2017, 9042851. SAS. 2012. Institute Inc. SAS/STAT Statistics user’s guide. Statistical analytical system, 5th rev ed. Cary, NC : SAS Institute Inc. Sheela, R.R., Babu, U., Mu, J., Elankumaran, S., Bautista, D.A., Raybourne, R.B. and et al. 2003. Immune responses against Salmonella enterica serovar Enteritidis infection in virally immunosuppressed chickens. Clin. Diagn. Lab. Immunol. 10, 670679. Sheena M. and Arthur G. 1978. A micro method for the estimation of killing and phagocytosis of Candida albicans by human leucocytes. J. Immunol. Methods 20, 43–52. Shen, X., Yin, L., Zhang, A., Zhao, R., Yin, D., Wang, J. et al. 2023. Prevalence and characterization of Salmonella isolated from chickens in Anhui, China. Pathogens 12, 465. Sornplang, P., Leelavatcharamas, V. and Soikum, C. 2015. Heterophil phagocytic activity stimulated by Lactobacillus salivarius L61 and L55 supplementation in broilers with Salmonella infection. Asian-Australas. J. Anim. Sci. 28, 1657–1661. Sreekantapuram, S., Berens, C., Barth, A.S., Methner, U. and Berndt, A. 2021. Interaction of Salmonella gallinarum and Salmonella enteritidis with peripheral leucocytes of hens with different laying performance. Vet. Res. 52, 123. Steel, R.G.D. and Torrie, J.H. 1980. Principles and procedures of Statistics, New York, NY: McGraw-Hill Book Company Inc. Tambekar, D., Dhanorkar, D., Gulhane, S., Khandelwal, V. and Dudhane, M. 2006. Antibacterial susceptibility of some urinary tract pathogens to commonly used antibiotics. African J. Biotechnol. 2006, 5. Tizard, I. 2002. The avian antibody response. Semin. Avian Exot. Pet Med. 11(1), 2–14. Van Immerseel, F., De Buck, J., De Smet, I., Mast, J., Haesebrouck, F. and Ducatelle, R. 2002. The effect of vaccination with a Salmonella enteritidis aroA mutant on early cellular responses in cecal lamina propria of newly hatched chickens. Vaccine 20, 3034–3041. Wibisono, F.M., Wibisono, F.J., Effendi, M.H., Plumeriastuti, H., Hidayatullah, A.R., Hartadi, E.B. and et al. 2020. A review of Salmonellosis on poultry farms: public health importance. Sys. Rev. Pharm. 11(9), 481–486. Withanage, G.S.K., Kaiser, P., Wigley, P., Powers, C., Mastroeni, P., Brooks, H. and et al. 2004. Rapid expression of chemokines and proinflammatory cytokines in newly hatched chickens infected with Salmonella enterica serovar Typhimurium. Infect. Immun. 72, 2152–2159. Yuan, J.S., Reed, A., Chen, F. and Stewart, C.N. 2006. Statistical analysis of real-time PCR data. BMC Bioinformatics. 7, 85. | ||
| How to Cite this Article |
| Pubmed Style Mosa YI, Gharib AA, Galil SYA, Ali AM, Abo-remela EM. Transcriptional responses of cytokines, immunoglobulin A, and nitric oxide genes in one day old chicks post Salmonella typhimurium infection: An experimental study. Open Vet. J.. 2024; 14((1) (Zagazig Veterinary Conference)): 200-213. doi:10.5455/OVJ.2024.v14.i1.18 Web Style Mosa YI, Gharib AA, Galil SYA, Ali AM, Abo-remela EM. Transcriptional responses of cytokines, immunoglobulin A, and nitric oxide genes in one day old chicks post Salmonella typhimurium infection: An experimental study. https://www.openveterinaryjournal.com/?mno=175636 [Access: January 24, 2026]. doi:10.5455/OVJ.2024.v14.i1.18 AMA (American Medical Association) Style Mosa YI, Gharib AA, Galil SYA, Ali AM, Abo-remela EM. Transcriptional responses of cytokines, immunoglobulin A, and nitric oxide genes in one day old chicks post Salmonella typhimurium infection: An experimental study. Open Vet. J.. 2024; 14((1) (Zagazig Veterinary Conference)): 200-213. doi:10.5455/OVJ.2024.v14.i1.18 Vancouver/ICMJE Style Mosa YI, Gharib AA, Galil SYA, Ali AM, Abo-remela EM. Transcriptional responses of cytokines, immunoglobulin A, and nitric oxide genes in one day old chicks post Salmonella typhimurium infection: An experimental study. Open Vet. J.. (2024), [cited January 24, 2026]; 14((1) (Zagazig Veterinary Conference)): 200-213. doi:10.5455/OVJ.2024.v14.i1.18 Harvard Style Mosa, Y. I., Gharib, . A. A., Galil, . S. Y. A., Ali, . A. M. & Abo-remela, . E. M. (2024) Transcriptional responses of cytokines, immunoglobulin A, and nitric oxide genes in one day old chicks post Salmonella typhimurium infection: An experimental study. Open Vet. J., 14 ((1) (Zagazig Veterinary Conference)), 200-213. doi:10.5455/OVJ.2024.v14.i1.18 Turabian Style Mosa, Yasmine I., Ahlam A. Gharib, Sara Y. Abd-el Galil, Amira. M. Ali, and Etab M. Abo-remela. 2024. Transcriptional responses of cytokines, immunoglobulin A, and nitric oxide genes in one day old chicks post Salmonella typhimurium infection: An experimental study. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 200-213. doi:10.5455/OVJ.2024.v14.i1.18 Chicago Style Mosa, Yasmine I., Ahlam A. Gharib, Sara Y. Abd-el Galil, Amira. M. Ali, and Etab M. Abo-remela. "Transcriptional responses of cytokines, immunoglobulin A, and nitric oxide genes in one day old chicks post Salmonella typhimurium infection: An experimental study." Open Veterinary Journal 14 (2024), 200-213. doi:10.5455/OVJ.2024.v14.i1.18 MLA (The Modern Language Association) Style Mosa, Yasmine I., Ahlam A. Gharib, Sara Y. Abd-el Galil, Amira. M. Ali, and Etab M. Abo-remela. "Transcriptional responses of cytokines, immunoglobulin A, and nitric oxide genes in one day old chicks post Salmonella typhimurium infection: An experimental study." Open Veterinary Journal 14.(1) (Zagazig Veterinary Conference) (2024), 200-213. Print. doi:10.5455/OVJ.2024.v14.i1.18 APA (American Psychological Association) Style Mosa, Y. I., Gharib, . A. A., Galil, . S. Y. A., Ali, . A. M. & Abo-remela, . E. M. (2024) Transcriptional responses of cytokines, immunoglobulin A, and nitric oxide genes in one day old chicks post Salmonella typhimurium infection: An experimental study. Open Veterinary Journal, 14 ((1) (Zagazig Veterinary Conference)), 200-213. doi:10.5455/OVJ.2024.v14.i1.18 |